Comparison of Adjuvanted-Whole Inactivated Virus and Live-Attenuated Virus Vaccines against Challenge with Contemporary, Antigenically Distinct H3N2 Influenza A Viruses
- PMID: 30185589
- PMCID: PMC6206469
- DOI: 10.1128/JVI.01323-18
Comparison of Adjuvanted-Whole Inactivated Virus and Live-Attenuated Virus Vaccines against Challenge with Contemporary, Antigenically Distinct H3N2 Influenza A Viruses
Abstract
Influenza A viruses in swine (IAV-S) circulating in the United States of America are phylogenetically and antigenically distinct. A human H3 hemagglutinin (HA) was introduced into the IAV-S gene pool in the late 1990s, sustained continued circulation, and evolved into five monophyletic genetic clades, H3 clades IV-A to -E, after 2009. Across these phylogenetic clades, distinct antigenic clusters were identified, with three clusters (cyan, red, and green antigenic cluster) among the most frequently detected antigenic phenotypes (Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, et al. J Virol 90:8266-8280, 2016, https://doi.org/10.1128/JVI.01002-16). Although it was demonstrated that antigenic diversity of H3N2 IAV-S was associated with changes at a few amino acid positions in the head of the HA, the implications of this diversity for vaccine efficacy were not tested. Using antigenically representative H3N2 viruses, we compared whole inactivated virus (WIV) and live-attenuated influenza virus (LAIV) vaccines for protection against challenge with antigenically distinct H3N2 viruses in pigs. WIV provided partial protection against antigenically distinct viruses but did not prevent virus replication in the upper respiratory tract. In contrast, LAIV provided complete protection from disease and virus was not detected after challenge with antigenically distinct viruses.IMPORTANCE Due to the rapid evolution of the influenza A virus, vaccines require continuous strain updates. Additionally, the platform used to deliver the vaccine can have an impact on the breadth of protection. Currently, there are various vaccine platforms available to prevent influenza A virus infection in swine, and we experimentally tested two: adjuvanted-whole inactivated virus and live-attenuated virus. When challenged with an antigenically distinct virus, adjuvanted-whole inactivated virus provided partial protection, while live-attenuated virus provided effective protection. Additional strategies are required to broaden the protective properties of inactivated virus vaccines, given the dynamic antigenic landscape of cocirculating strains in North America, whereas live-attenuated vaccines may require less frequent strain updates, based on demonstrated cross-protection. Enhancing vaccine efficacy to control influenza infections in swine will help reduce the impact they have on swine production and reduce the risk of swine-to-human transmission.
Keywords: influenza; live-attenuated; swine; vaccine.
Figures
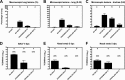
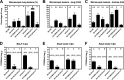
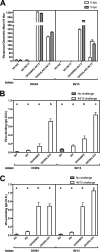
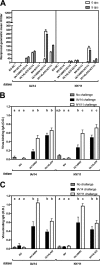
References
-
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

