A Proteomic Atlas of the African Swine Fever Virus Particle
- PMID: 30185597
- PMCID: PMC6232493
- DOI: 10.1128/JVI.01293-18
A Proteomic Atlas of the African Swine Fever Virus Particle
Abstract
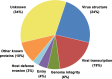
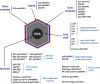
African swine fever virus (ASFV) is a large and complex DNA virus that causes a highly lethal swine disease for which there is no vaccine available. The ASFV particle, with an icosahedral multilayered structure, contains multiple polypeptides whose identity is largely unknown. Here, we analyzed by mass spectroscopy the protein composition of highly purified extracellular ASFV particles and performed immunoelectron microscopy to localize several of the detected proteins. The proteomic analysis identified 68 viral proteins, which account for 39% of the genome coding capacity. The ASFV proteome includes essentially all the previously described virion proteins and, interestingly, 44 newly identified virus-packaged polypeptides, half of which have an unknown function. A great proportion of the virion proteins are committed to the virus architecture, including two newly identified structural proteins, p5 and p8, which are derived from the core polyproteins pp220 and pp62, respectively. In addition, the virion contains a full complement of enzymes and factors involved in viral transcription, various enzymes implicated in DNA repair and protein modification, and some proteins concerned with virus entry and host defense evasion. Finally, 21 host proteins, many of them localized at the cell surface and related to the cortical actin cytoskeleton, were reproducibly detected in the ASFV particle. Immunoelectron microscopy strongly supports the suggestion that these host membrane-associated proteins are recruited during virus budding at actin-dependent membrane protrusions. Altogether, the results of this study provide a comprehensive model of the ASFV architecture that integrates both compositional and structural information.IMPORTANCE African swine fever virus causes a highly contagious and lethal disease of swine that currently affects many countries of sub-Saharan Africa, the Caucasus, the Russian Federation, and Eastern Europe and has very recently spread to China. Despite extensive research, effective vaccines or antiviral strategies are still lacking, and many basic questions on the molecular mechanisms underlying the infective cycle remain. One such gap regards the composition and structure of the infectious virus particle. In the study described in this report, we identified the set of viral and host proteins that compose the virion and determined or inferred the localization of many of them. This information significantly increases our understanding of the biological and structural features of an infectious African swine fever virus particle and will help direct future research efforts.
Keywords: African swine fever virus; NCLDV; giant virus; immunoelectron microscopy; mass spectrometry; proteome; proteomic analysis; virus composition; virus structure.
Copyright © 2018 American Society for Microbiology.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

