Structural insights into the functional diversity of the CDK-cyclin family
- PMID: 30185601
- PMCID: PMC6170502
- DOI: 10.1098/rsob.180112
Structural insights into the functional diversity of the CDK-cyclin family
Abstract
Since their characterization as conserved modules that regulate progression through the eukaryotic cell cycle, cyclin-dependent protein kinases (CDKs) in higher eukaryotic cells are now also emerging as significant regulators of transcription, metabolism and cell differentiation. The cyclins, though originally characterized as CDK partners, also have CDK-independent roles that include the regulation of DNA damage repair and transcriptional programmes that direct cell differentiation, apoptosis and metabolic flux. This review compares the structures of the members of the CDK and cyclin families determined by X-ray crystallography, and considers what mechanistic insights they provide to guide functional studies and distinguish CDK- and cyclin-specific activities. Aberrant CDK activity is a hallmark of a number of diseases, and structural studies can provide important insights to identify novel routes to therapy.
Keywords: cell cycle; cyclin; kinase; transcription.
© 2018 The Authors.
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript. Some work in the authors' laboratory is funded by Astex Pharmaceuticals.
Figures

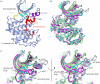
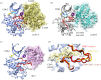
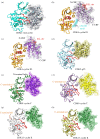
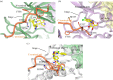




References
-
- Morgan DO. 2007. The cell cycle principles of control (primers in biology). London, UK: New Science Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
