Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis
- PMID: 30185668
- PMCID: PMC6171814
- DOI: 10.1172/jci.insight.122061
Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis
Abstract
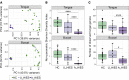
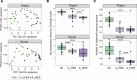
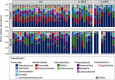
Studies in patients with genetic defects can provide unique insights regarding the role of specific genes and pathways in humans. Patients with defects in the Th17/IL-17 axis, such as patients harboring loss-of-function STAT3 mutations (autosomal-dominant hyper IgE syndrome; AD-HIES) present with recurrent oral fungal infections. Our studies aimed to comprehensively evaluate consequences of STAT3 deficiency on the oral commensal microbiome. We characterized fungal and bacterial communities in AD-HIES in the presence and absence of oral fungal infection compared with healthy volunteers. Analyses of oral mucosal fungal communities in AD-HIES revealed severe dysbiosis with dominance of Candida albicans (C. albicans) in actively infected patients and minimal representation of health-associated fungi and/or opportunists. Bacterial communities also displayed dysbiosis in AD-HIES, particularly in the setting of active Candida infection. Active candidiasis was associated with decreased microbial diversity and enrichment of the streptococci Streptococcus oralis (S. oralis) and S. mutans, suggesting an interkingdom interaction of C. albicans with oral streptococci. Increased abundance of S. mutans was consistent with susceptibility to dental caries in AD-HIES. Collectively, our findings illustrate a critical role for STAT3/Th17 in the containment of C. albicans as a commensal organism and an overall contribution in the establishment of fungal and bacterial oral commensal communities.
Keywords: Fungal infections; Infectious disease; Microbiology; Monogenic diseases.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

