MEG3 is increased in idiopathic pulmonary fibrosis and regulates epithelial cell differentiation
- PMID: 30185671
- PMCID: PMC6171798
- DOI: 10.1172/jci.insight.122490
MEG3 is increased in idiopathic pulmonary fibrosis and regulates epithelial cell differentiation
Abstract
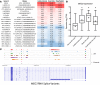
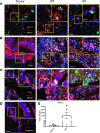
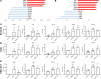
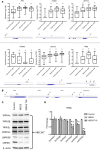
Idiopathic pulmonary fibrosis (IPF) is a chronic interstitial lung disease causing fibrotic remodeling of the peripheral lung, leading to respiratory failure. Peripheral pulmonary epithelial cells lose normal alveolar epithelial gene expression patterns and variably express genes associated with diverse conducting airway epithelial cells, including basal cells. Single-cell RNA sequencing of pulmonary epithelial cells isolated from IPF lung tissue demonstrated altered expression of LncRNAs, including increased MEG3. MEG3 RNA was highly expressed in subsets of the atypical IPF epithelial cells and correlated with conducting airway epithelial gene expression patterns. Expression of MEG3 in human pulmonary epithelial cell lines increased basal cell-associated RNAs, including TP63, KRT14, STAT3, and YAP1, and enhanced cell migration, consistent with a role for MEG3 in regulating basal cell identity. MEG3 reduced expression of TP73, SOX2, and Notch-associated RNAs HES1 and HEY1, in primary human bronchial epithelial cells, demonstrating a role for MEG3 in the inhibition of genes influencing basal cell differentiation into club, ciliated, or goblet cells. MEG3 induced basal cell genes and suppressed genes associated with terminal differentiation of airway cells, supporting a role for MEG3 in regulation of basal progenitor cell functions, which may contribute to tissue remodeling in IPF.
Keywords: Fibrosis; Noncoding RNAs; Pulmonology.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

