New Insights Into Wall Polysaccharide O-Acetylation
- PMID: 30186297
- PMCID: PMC6110886
- DOI: 10.3389/fpls.2018.01210
New Insights Into Wall Polysaccharide O-Acetylation
Abstract
The extracellular matrix of plants, algae, bacteria, fungi, and some archaea consist of a semipermeable composite containing polysaccharides. Many of these polysaccharides are O-acetylated imparting important physiochemical properties to the polymers. The position and degree of O-acetylation is genetically determined and varies between organisms, cell types, and developmental stages. Despite the importance of wall polysaccharide O-acetylation, only recently progress has been made to elucidate the molecular mechanism of O-acetylation. In plants, three protein families are involved in the transfer of the acetyl substituents to the various polysaccharides. In other organisms, this mechanism seems to be conserved, although the number of required components varies. In this review, we provide an update on the latest advances on plant polysaccharide O-acetylation and related information from other wall polysaccharide O-acetylating organisms such as bacteria and fungi. The biotechnological impact of understanding wall polysaccharide O-acetylation ranges from the design of novel drugs against human pathogenic bacteria to the development of improved lignocellulosic feedstocks for biofuel production.
Keywords: O-acetylation; biosynthesis; cell wall; mechanism; polysaccharides.
Figures
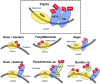
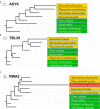
References
-
- Baker P., Ricer T., Moynihan P. J., Kitova E. N., Walvoort M. T., Little D. J., et al. (2014). P. aeruginosa SGNH hydrolase-like proteins AlgJ and AlgX have similar topology but separate and distinct roles in alginate acetylation. PLoS Pathog. 10:e1004334. 10.1371/journal.ppat.1004334 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

