Integrated analysis of key mRNAs and lncRNAs in osteoarthritis
- PMID: 30186409
- PMCID: PMC6122320
- DOI: 10.3892/etm.2018.6360
Integrated analysis of key mRNAs and lncRNAs in osteoarthritis
Abstract
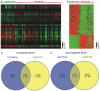
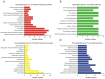
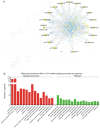
Osteoarthritis (OA) is the most common type of arthritis, observed mainly in the population aged >65 years. However, the mechanism underlying the development and progression of OA has remained largely elusive. The present study aimed to identify differentially expressed mRNAs and lncRNAs in OA. By analyzing the GSE48556 and GSE82107 datasets, a total of 202 up- and 434 downregulated mRNAs were identified in OA. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis indicated that differently expressed genes were mainly involved in regulating antigen processing and presentation, interspecies interaction between organisms, immune response, transcription and signal transduction. In addition, a series of long non-coding (lnc)RNAs were differently expressed in OA. To provide novel data on the molecular mechanisms and functional roles of these lncRNAs in OA, a co-expression analysis was performed, which revealed that the dysregulated lncRNAs were associated with transcription, signal transduction, immune response and cell adhesion. In addition, certain key genes in protein-protein interaction networks were identified. The present study provided useful information for exploring potential candidate biomarkers for the diagnosis and prognosis of OA, as well as novel drug targets.
Keywords: differentially expressed gene; expression profiling; long non-coding RNA; osteoarthritis; protein-protein interaction network.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
