2D DIGE proteomic analysis reveals fasting-induced protein remodeling through organ-specific transcription factor(s) in mice
- PMID: 30186752
- PMCID: PMC6120221
- DOI: 10.1002/2211-5463.12497
2D DIGE proteomic analysis reveals fasting-induced protein remodeling through organ-specific transcription factor(s) in mice
Abstract
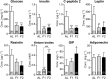
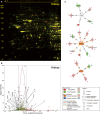
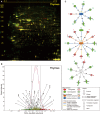
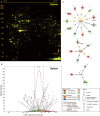
Overnight fasting is a routine procedure before surgery in clinical settings. Intermittent fasting is the most common diet/fitness trend implemented for weight loss and the treatment of lifestyle-related diseases. In either setting, the effects not directly related to parameters of interest, either beneficial or harmful, are often ignored. We previously demonstrated differential activation of cellular adaptive responses in 13 atrophied/nonatrophied organs of fasted mice by quantitative PCR analysis of gene expression. Here, we investigated 2-day fasting-induced protein remodeling in six major mouse organs (liver, kidney, thymus, spleen, brain, and testis) using two-dimensional difference gel electrophoresis (2D DIGE) proteomics as an alternative means to examine systemic adaptive responses. Quantitative analysis of protein expression followed by protein identification using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOFMS) revealed that the expression levels of 72, 26, and 14 proteins were significantly up- or downregulated in the highly atrophied liver, thymus, and spleen, respectively, and the expression levels of 32 proteins were up- or downregulated in the mildly atrophied kidney. Conversely, there were no significant protein expression changes in the nonatrophied organs, brain and testis. Upstream regulator analysis highlighted transcriptional regulation by peroxisome proliferator-activated receptor alpha (PPARα) in the liver and kidney and by tumor protein/suppressor p53 (TP53) in the thymus, spleen, and liver. These results imply of the existence of both common and distinct adaptive responses between major mouse organs, which involve transcriptional regulation of specific protein expression upon short-term fasting. Our data may be valuable in understanding systemic transcriptional regulation upon fasting in experimental animals.
Keywords: 2D DIGE; PPARα; TP53; fasting; proteomics; transcriptional regulation.
Figures


References
-
- Jensen TL, Kiersgaard MK, Sorensen DB and Mikkelsen LF (2013) Fasting of mice: a review. Lab Anim 47, 225–240. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

