Rapid, field-deployable method for collecting and preserving plant metabolome for biochemical and functional characterization
- PMID: 30188945
- PMCID: PMC6126852
- DOI: 10.1371/journal.pone.0203569
Rapid, field-deployable method for collecting and preserving plant metabolome for biochemical and functional characterization
Abstract
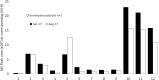
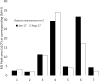
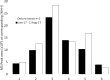
Study of plant metabolome is a growing field of science that catalogs vast biochemical and functional diversity of phytochemicals. However, collecting and storing samples of plant metabolome, sharing these samples across the scientific community and making them compatible with bioactivity assays presents significant challenges to the advancement of metabolome research. We have developed a RApid Metabolome Extraction and Storage (RAMES) technology that allows efficient, highly compact, field-deployable collection and storage of libraries of plant metabolome. RAMES technology combines rapid extraction with immobilization of extracts on glass microfiber filter discs. Two grams of plant tissue extracted in ethanol, using a specially adapted Dremel® rotary tool, produces 25-35 replicas of 10 mm glass fiber discs impregnated with phytochemicals. These discs can be either eluted with solvents (such as 70% ethanol) to study the metabolomic profiles or used directly in a variety of functional assays. We have developed simple, non-sterile, anti-fungal, anti-bacterial, and anti-oxidant assays formatted for 24-multiwell plates directly compatible with RAMES discs placed inside the wells. Using these methods we confirmed activity in 30 out of 32 randomly selected anti-microbial medicinal plants and spices. Seven species scored the highest activity (total kill) in the anti-bacterial (bacteria from human saliva) and two anti-fungal screens (Fusarium spp. and Saccharomyces cerevisiae), providing functional validation of RAMES technology. RAMES libraries showed limited degradation of compounds after 12 months of storage at -20°C, while others remained stable. Fifty-eight percent of structures characterized in the extracts loaded onto RAMES discs could be eluted from the discs without significant losses. Miniaturized RAMES technology, as described and validated in this manuscript offers a labor, cost, and time-effective alternative to conventional collection of phytochemicals. RAMES technology enables creation of comprehensive metabolomic libraries from various ecosystems and geographical regions in a format compatible with further biochemical and functional studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Dixon RA. Natural products and plant disease resistance. Nature [Internet]. 2001;411(6839):843–7. Available from: http://www.nature.com/doifinder/10.1038/35081178 - DOI - PubMed
-
- Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG. Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci [Internet]. 2017;114(22):5601–6. Available from: http://www.pnas.org/lookup/doi/10.1073/pnas.1614680114 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

