Whole Genome Analysis of Cyclin Dependent Kinase (CDK) Gene Family in Cotton and Functional Evaluation of the Role of CDKF4 Gene in Drought and Salt Stress Tolerance in Plants
- PMID: 30189594
- PMCID: PMC6164816
- DOI: 10.3390/ijms19092625
Whole Genome Analysis of Cyclin Dependent Kinase (CDK) Gene Family in Cotton and Functional Evaluation of the Role of CDKF4 Gene in Drought and Salt Stress Tolerance in Plants
Abstract
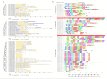
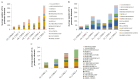
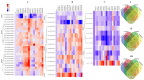
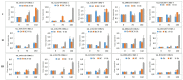
Cotton (Gossypium spp.) is the number one crop cultivated for fiber production and the cornerstone of the textile industry. Drought and salt stress are the major abiotic stresses, which can have a huge economic impact on cotton production; this has been aggravated with continued climate change, and compounded by pollution. Various survival strategies evolved by plants include the induction of various stress responsive genes, such as cyclin dependent kinases (CDKs). In this study, we performed a whole-genome identification and analysis of the CDK gene family in cotton. We identified 31, 12, and 15 CDK genes in G. hirsutum, G. arboreum, and G. raimondii respectively, and they were classified into 6 groups. CDK genes were distributed in 15, 10, and 9 linkage groups of AD, D, and A genomes, respectively. Evolutionary analysis revealed that segmental types of gene duplication were the primary force underlying CDK genes expansion. RNA sequence and RT-qPCR validation revealed that Gh_D12G2017 (CDKF4) was strongly induced by drought and salt stresses. The transient expression of Gh_D12G2017-GFP fusion protein in the protoplast showed that Gh_D12G2017 was localized in the nucleus. The transgenic Arabidopsis lines exhibited higher concentration levels of the antioxidant enzymes measured, including peroxidase (POD), superoxide dismutase (SOD), and catalase (CAT) concentrations under drought and salt stress conditions with very low levels of oxidants. Moreover, cell membrane stability (CMS), excised leaf water loss (ELWL), saturated leaf weight (SLW), and chlorophyll content measurements showed that the transgenic Arabidopsis lines were highly tolerant to either of the stress factors compared to their wild types. Moreover, the expression of the stress-related genes was also significantly up-regulated in Gh_D12G2017(CDKF4) transgenic Arabidopsis plants under drought and salt conditions. We infer that CDKF-4s and CDKG-2s might be the primary regulators of salt and drought responses in cotton.
Keywords: abiotic stress; cotton cyclin dependent kinase; gene expression; oxidant and antioxidant enzymes; plant growth and development; segmental duplication; transgenic arabidopsis lines.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton.BMC Genet. 2019 Jul 23;20(1):62. doi: 10.1186/s12863-019-0756-6. BMC Genet. 2019. PMID: 31337336 Free PMC article.
-
Cotton Late Embryogenesis Abundant (LEA2) Genes Promote Root Growth and Confer Drought Stress Tolerance in Transgenic Arabidopsis thaliana.G3 (Bethesda). 2018 Jul 31;8(8):2781-2803. doi: 10.1534/g3.118.200423. G3 (Bethesda). 2018. PMID: 29934376 Free PMC article.
-
Genome-Wide Analysis of Multidrug and Toxic Compound Extrusion (MATE) Family in Gossypium raimondii and Gossypium arboreum and Its Expression Analysis Under Salt, Cadmium, and Drought Stress.G3 (Bethesda). 2018 Jul 2;8(7):2483-2500. doi: 10.1534/g3.118.200232. G3 (Bethesda). 2018. PMID: 29794162 Free PMC article.
-
Genome-wide expression analysis of phospholipase A1 (PLA1) gene family suggests phospholipase A1-32 gene responding to abiotic stresses in cotton.Int J Biol Macromol. 2021 Dec 1;192:1058-1074. doi: 10.1016/j.ijbiomac.2021.10.038. Epub 2021 Oct 14. Int J Biol Macromol. 2021. PMID: 34656543 Review.
-
Insights into Drought Stress Signaling in Plants and the Molecular Genetic Basis of Cotton Drought Tolerance.Cells. 2019 Dec 31;9(1):105. doi: 10.3390/cells9010105. Cells. 2019. PMID: 31906215 Free PMC article. Review.
Cited by
-
RNA-Sequencing, Physiological and RNAi Analyses Provide Insights into the Response Mechanism of the ABC-Mediated Resistance to Verticillium dahliae Infection in Cotton.Genes (Basel). 2019 Feb 1;10(2):110. doi: 10.3390/genes10020110. Genes (Basel). 2019. PMID: 30717226 Free PMC article.
-
Genome-wide analysis of the CalS gene family in cotton reveals their potential roles in fiber development and responses to stress.PeerJ. 2021 Nov 30;9:e12557. doi: 10.7717/peerj.12557. eCollection 2021. PeerJ. 2021. PMID: 34909280 Free PMC article.
-
SSR-Linkage map of interspecific populations derived from Gossypium trilobum and Gossypium thurberi and determination of genes harbored within the segregating distortion regions.PLoS One. 2018 Nov 12;13(11):e0207271. doi: 10.1371/journal.pone.0207271. eCollection 2018. PLoS One. 2018. PMID: 30419064 Free PMC article.
-
Analysis of Whole-Transcriptome RNA-Seq Data Reveals the Involvement of Alternative Splicing in the Drought Response of Glycyrrhiza uralensis.Front Genet. 2022 May 17;13:885651. doi: 10.3389/fgene.2022.885651. eCollection 2022. Front Genet. 2022. PMID: 35656323 Free PMC article.
-
Genome-wide identification and analysis of anthocyanin synthesis-related R2R3-MYB genes in Cymbidium goeringii.Front Plant Sci. 2022 Sep 28;13:1002043. doi: 10.3389/fpls.2022.1002043. eCollection 2022. Front Plant Sci. 2022. PMID: 36247626 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

