Human Alzheimer's disease gene expression signatures and immune profile in APP mouse models: a discrete transcriptomic view of Aβ plaque pathology
- PMID: 30189875
- PMCID: PMC6127905
- DOI: 10.1186/s12974-018-1265-7
Human Alzheimer's disease gene expression signatures and immune profile in APP mouse models: a discrete transcriptomic view of Aβ plaque pathology
Abstract
Background: Alzheimer's disease (AD) is a chronic neurodegenerative disease with pathological hallmarks including the formation of extracellular aggregates of amyloid-beta (Aβ) known as plaques and intracellular tau tangles. Coincident with the formation of Aβ plaques is recruitment and activation of glial cells to the plaque forming a plaque niche. In addition to histological data showing the formation of the niche, AD genetic studies have added to the growing appreciation of how dysfunctional glia pathways drive neuropathology, with emphasis on microglia pathways. Genomic approaches enable comparisons of human disease profiles between different mouse models informing on their utility to evaluate secondary changes to triggers such as Aβ deposition.
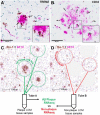
Methods: In this study, we utilized two animal models of AD to examine and characterize the AD-associated pathology: the Tg2576 Swedish APP (KM670/671NL) and TgCRND8 Swedish plus Indiana APP (KM670/671NL + V717F) lines. We used laser capture microscopy (LCM) to isolate samples surrounding Thio-S positive plaques from distal non-plaque tissue. These samples were then analyzed using RNA sequencing.
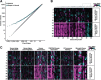
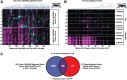
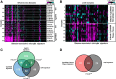
Results: We determined age-associated transcriptomic differences between two similar yet distinct APP transgenic mouse models, known to differ in proportional amyloidogenic species and plaque deposition rates. In Tg2576, human AD gene signatures were not observed despite profiling mice out to 15 months of age. TgCRND8 mice however showed progressive and robust induction of lysomal, neuroimmune, and ITIM/ITAM-associated gene signatures overlapping with prior human AD brain transcriptomic studies. Notably, RNAseq analyses highlighted the vast majority of transcriptional changes observed in aging TgCRND8 cortical brain homogenates were in fact specifically enriched within the plaque niche samples. Data uncovered plaque-associated enrichment of microglia-related genes such as ITIM/ITAM-associated genes and pathway markers of phagocytosis.
Conclusion: This work may help guide improved translational value of APP mouse models of AD, particularly for strategies aimed at targeting neuroimmune and neurodegenerative pathways, by demonstrating that TgCRND8 more closely recapitulates specific human AD-associated transcriptional responses.
Keywords: Alzheimer’s disease; Microglia; Neuroinflammation; Plaque; Transcriptomics.
Conflict of interest statement
Ethics approval and consent to participate
Principles of laboratory animal care were followed, and all studies were previously approved by the Institutional Animal Care and Use Committee and were performed in accordance to the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the National Institutes of Health (Library of Congress Control Number 2010940400, revised 2011).
Consent for publication
Not applicable
Competing interests
All authors were or are employees and shareholders in Merck & Co., Inc. at the time of their contributions. The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

