Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors
- PMID: 30190312
- PMCID: PMC6168235
- DOI: 10.1085/jgp.201711979
Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors
Erratum in
-
Correction: Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors.J Gen Physiol. 2025 Jan 6;157(1):e20171197912022024c. doi: 10.1085/jgp.20171197912022024c. Epub 2024 Dec 19. J Gen Physiol. 2025. PMID: 39699937 Free PMC article. No abstract available.
Abstract
Recent breakthroughs and developments in structural biology have led to a spate of crystal structures for G protein-coupled receptors (GPCRs). This is the case for the muscarinic acetylcholine receptors (mAChRs) where inactive-state structures for four of the five subtypes and two active-state structures for one subtype are available. These mAChR crystal structures have provided new insights into receptor mechanisms, dynamics, and allosteric modulation. This is highly relevant to the mAChRs given that these receptors are an exemplar model system for the study of GPCR allostery. Allosteric mechanisms of the mAChRs are predominantly consistent with a two-state model, albeit with some notable recent exceptions. Herein, we discuss the mechanisms for positive and negative allosteric modulation at the mAChRs and compare and contrast these to evidence offered by pharmacological, biochemical, and computational approaches. This analysis provides insight into the fundamental pharmacological properties exhibited by GPCR allosteric modulators, such as enhanced subtype selectivity, probe dependence, and biased modulation while highlighting the current challenges that remain. Though complex, enhanced molecular understanding of allosteric mechanisms will have considerable influence on our understanding of GPCR activation and signaling and development of therapeutic interventions.
© 2018 Burger et al.
Figures

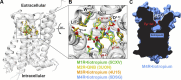





![Figure 9. Positive allosteric modulation of monomeric M4 mAChRs. (A
and B) [3H]NMS interaction binding studies between the PAM, LY2033298, and
acetylcholine at the M4 mAChR expressed in either CHO cells (A; replotted
from Leach et al., 2010) or purified from Sf9 cells (B; M4R-mT4L; for
methods, see Thal et al., 2016) and reconstituted into rHDL particles, as
was previously done for rhodopsin (Whorton et al., 2007). Nearly identical
levels of cooperativity were observed, indicating that LY2033298 is able to
influence the binding of acetylcholine in a monomeric mAChR and is not
dependent on oligomerization status of the receptor. Data from B represent
the mean ± SEM of n = 3 experiments performed in duplicate.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ea27/6168235/f02b15af6d45/jgp_201711979_fig9.gif)
References
-
- Ballesteros, J.A., and Weinstein H.. 1995. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neurosciences. 366–428. 10.1016/S1043-9471(05)80049-7 - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

