Selective depletion of metastatic stem cells as therapy for human colorectal cancer
- PMID: 30190334
- PMCID: PMC6180303
- DOI: 10.15252/emmm.201708772
Selective depletion of metastatic stem cells as therapy for human colorectal cancer
Abstract
Selective elimination of metastatic stem cells (MetSCs) promises to block metastatic dissemination. Colorectal cancer (CRC) cells overexpressing CXCR4 display trafficking functions and metastasis-initiating capacity. We assessed the antimetastatic activity of a nanoconjugate (T22-GFP-H6-FdU) that selectively delivers Floxuridine to CXCR4+ cells. In contrast to free oligo-FdU, intravenous T22-GFP-H6-FdU selectively accumulates and internalizes in CXCR4+ cancer cells, triggering DNA damage and apoptosis, which leads to their selective elimination and to reduced tumor re-initiation capacity. Repeated T22-GFP-H6-FdU administration in cell line and patient-derived CRC models blocks intravasation and completely prevents metastases development in 38-83% of mice, while showing CXCR4 expression-dependent and site-dependent reduction in foci number and size in liver, peritoneal, or lung metastases in the rest of mice, compared to free oligo-FdU. T22-GFP-H6-FdU induces also higher regression of established metastases than free oligo-FdU, with negligible distribution or toxicity in normal tissues. This targeted drug delivery approach yields potent antimetastatic effect, through selective depletion of metastatic CXCR4+ cancer cells, and validates metastatic stem cells (MetSCs) as targets for clinical therapy.
Keywords: CXCR4 receptor; colorectal cancer; metastatic stem cells; protein nanoconjugate; targeted drug delivery.
© 2018 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

- A
The nanoconjugate contains a fusion protein [T22‐GFP‐H6—composed of the peptide T22 as a CXCR4 ligand, a green fluorescent protein and a histidine tail—bound to the payload drug (Unzueta et al, 2012a)].
- B
Three to four pentameric oligonucleotides (approximately 20 molecules) of the antitumor drug 5‐Fluoro‐2′‐deoxyuridine (FdU), named oligo‐FdU, are conjugated to the T22‐GFP‐H6 targeting vector using a linker.
- C
T22‐GFP‐H6‐FdU chemical synthesis: T22‐GFP‐H6 is first covalently bound to the 6‐Maleimidohexanoic acid N‐hydroxysuccinimide ester linker through its amino groups in the external lysines (Hermanson, 2013). The thiol‐functionalized oligo‐FdU (oligo‐(FdU)5‐SH; see Appendix Fig S1) is then reacted with T22‐GFP‐H6 functionalized with maleimide (Michael reaction).
- D
High and constitutive expression of CXCR4 in the membrane of SW1417 CRC cells as measured by flow cytometry.
- E
Lack of human SDF‐1α release from cultured SW1417 CRC cells, as measured by ELISA, whereas human control 1BR3.G fibroblasts express high SDF‐1α levels, after 48 or 72 h of growth in culture (mean ± s.e.m., N = 2 experiment in duplicate).
- F
Nanoconjugate internalization in CXCR4‐overexpressing (CXCR4+) SW1417 CRC cells after 1‐h exposure at 1 μM, as measured by fluorescence emission using flow cytometry (mean ± s.e.m., N = 3 experiments in duplicate). Significant difference at **P = 0.002 between the T22‐GFP‐H6‐FdU and the T22‐GFP‐H6‐FdU + AMD3100 groups, Mann–Whitney U‐test.
- G
Intracellular trafficking of T22‐GFP‐H6‐FdU in CXCR4+ SW1417 cells by confocal microscopy after exposure at 1 μM for 24 h. The green staining corresponds to GFP‐containing nanoconjugates, and the red staining corresponds to plasma cell membranes stained with a red dye (CellMask™), whereas cell nucleus was stained in blue with Hoechst. The insets show detail of the intracellular localization of nanostructured, fluorescent entities, in an isosurface representation within a three‐dimensional volumetric x‐y‐z data field.
- H
Linearized T22‐GFP‐H6‐FdU dose–response trend line representation compared with unconjugated free oligo‐FdU exposure. Antitumor effect was measured as CXCR4+ SW1417 cell viability by MTT after 72‐h exposure as the described concentrations (mean ± s.e.m., N = 3 experiments in duplicate).
- I
Reduction in cell viability determined by optical microscope images of SW1417 cells exposed to 1 μM T22‐GFP‐H6‐FdU for 72 h, as compared to T22‐GFP‐H6 or free oligo‐FdU (N = 3 experiments in duplicate; Scale bar, 100 μm).

- A
Approach to achieve targeted drug delivery and selective killing of metastatic stem cells: CXCR4‐nanoconjugate interaction triggers CXCR4‐mediated internalization in MetSCs, in primary tumors and metastatic foci, followed by FdU release to the cytosol and diffusion to the nucleus to induce double‐strand breaks leading to selective killing of CXCR4+ cells.
- B
Selective T22‐GFP‐H6‐FdU nanoconjugate biodistribution in subcutaneous CXCR4+ SW1417 CRC tumor tissue 5 h after a 100 μg single intravenous dose, as measured by fluorescence emission using IVIS Spectrum 200 (N = 5/group). Biodistribution is similar to that achieved by the T22‐GFP‐H6 targeting vector and undetectable after Buffer or free oligo‐FdU treatment (N = 5 mice/group).
- C
Co‐localization (yellow merged) of the T22‐GFP‐H6‐FdU (green) and the CXCR4 receptor (red) and release of T22‐GFP‐H6‐FdU into the cytosol in CXCR4+ tumor cells 5 h after a 100 μg dose of nanoconjugate, as measured by dual anti‐GFP/anti‐CXCR4 immunofluorescence (IF). DAPI (blue nuclear staining). Fluorescence emission was measured in the green and red channels using the ImageJ software and expressed as mean area (A) ± s.e.m (μm2) (N = 10, 2 tumor fields × 5 mice; 200×). Note the significant (P = 0.003) increase in the area occupied by the green dots (nanoconjugate released to the cell cytosol) in T22‐GFP‐H6‐FdU‐treated tumors, compared to free oligo‐FdU‐treated control tissues. Scale bar, 50 μm.
- D
Administration of the CXCR4 antagonist AMD3100 completely blocks T22‐GFP‐H6‐FdU tumor biodistribution, as measured by fluorescence emission. Fluorescence is not detected in Buffer or free oligo‐FdU controls (N = 5 tumor fields/group).
- E
The uptake of T22‐GFP‐H6‐FdU observed in CXCR4+ SW1417 tumor tissues is almost completely blocked by prior AMD3100 administration, as quantified using the anti‐GFP IHC H‐score (mean ± s.e.m., N = 5 tumor fields/group). Comparison of T22‐GFP‐H6 uptake between groups: (B: Buffer; F: free oligo‐FdU; T‐F: T22‐GFP‐H6‐FdU; T‐F+A: T22‐GFP‐H6‐FdU+AMD3100). P‐values for statistical differences B vs. T‐F, **P = 0.000; F vs. T‐F, **P = 0.000; T‐F vs. TFA, **P = 0.004. Mann–Whitney U‐test.
- F
Representative images of T22‐GFP‐H6‐FdU uptake and AMD3100 competition by anti‐GFP immunostaining, which quantitation is reported in panel (E). Scale bar, 50 μm.
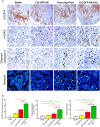
- A
Representative images of CXCR4 overexpression in subcutaneous tumor tissue, showing similar CXCR4 levels among compared groups (N = 5/group; Buffer, T22‐GFP‐H6‐FdU, T22‐GFP‐H6, and free oligo‐FdU) before treatment (upper panels). Representative images of DNA double‐strand break induction and caspase‐3 activation (measured with anti‐γ‐H2AX or anticleaved caspase‐3 by IHC) 5 h post‐administration (middle panels). Apoptotic induction (Hoechst staining, 24 h post‐administration, lower panels). Note the higher number of cells positive for DSBs, caspase‐3 activation, and apoptosis induction in the T22‐GFP‐H6‐FdU as compared to free oligo‐FdU. Black or white arrows indicate dead cells. Scale bar, 50 μm.
- B
Quantitation of the number of cells containing DSBs or active caspase‐3 in IHC‐stained tumor sections 5 h post‐treatment and the number of condensated or disaggregated nuclei (by Hoechst staining) 24 h post‐treatment in tumor sections of 10 high‐power fields (400× magnification) using the Cell∧D software (N = 50; 10 tumor fields/mice; 5 mice/group). Data expressed as mean ± s.e.m. Parameter comparison between groups: (B: Buffer; T: T22‐GFP‐H6; F: free oligo‐FdU; T‐F: T22‐GFP‐H6‐FdU). P‐values for statistical differences: γ‐H2AX staining quantitation: B vs. T, # P = 0.001; B vs. F, # P = 0.000; B vs. T‐F, # P = 0.000; T vs. T‐F, **P = 0.001; F vs. T‐F, *P = 0.02. Cleaved caspase‐3 quantitation: B vs. F, *P = 0.034; B vs. T‐F, **P = 0.009; T vs. T‐F, **P = 0.003; F vs. T‐F, *P = 0.012. Hoechst staining quantitation: B vs. F, **P = 0.01; B vs. T‐F, **P = 0.001; T vs. T‐F, **P = 0.000; F vs. T‐F, *P = 0.032. Mann Whitney U‐test.

- A, B
T22‐GFP‐H6‐FdU depletes CXCR4+ cancer cells from SW1417 CRC tumor tissue after a 100 μg single‐dose administration. Note the reduction in CXCR4+ cell fraction in the tumor 24 h after injection, their almost complete elimination at 48 h, and the re‐emergence of CXCR4+ cells 72 h post‐administration, using anti‐CXCR4 IHC. In contrast, the CXCR4+ cancer cell fraction (CXCR4+ CCF) in tumor tissue remains constant along time after free oligo‐FdU treatment. The 3‐day time‐lapse for CXCR4+ tumor cell re‐appearance defines the dosage interval used in a repeated dose schedule of nanoconjugate administration in the experiments to evaluate its antimetastatic effect (N = 5: 5 mice/group; 1 samples/mouse). Scale bar, 50 μm. Data expressed as mean ± s.e.m. CXCR4 H‐score comparison for T22‐GFP‐H6‐FdU(T‐F)‐treated tumors among time points (green line, panel A). P‐values for statistical differences: T‐F Basal vs. T‐F 24 h, *P = 0.038; T‐F Basal vs. T‐F 48 h, **P = 0.001; T‐F Basal vs. T‐F 72 h, **P = 0.003; T‐F 24 h vs. T‐F 48 h, *P = 0.033. CXCR4 H‐score comparison between T22‐GFP‐H6‐FdU (T‐F) and free oligo‐FdU (F) (black line, panel A). P‐values for statistical differences: T‐F vs. F at 48 h, **P = 0.001; T‐F vs. F at 72 h, *P = 0.034). Mann–Whitney U‐test.
- C, D
Significant reduction in the number of spheroid formed (C, optical microscope) and their bioluminescence emission (D, IVIS Spectrum 200), generated by 1 × 106 disaggregated cells (cultured in stem cell‐conditioned media and low‐adhesion plates), obtained from CXCR4+ luciferase+ SW1417 subcutaneous tumors, 24 h after 100 μg T22‐GFP‐H6‐FdU intravenous doses, for 2 consecutive days, as compared to Buffer‐treated or free oligo‐FdU‐treated mice. (D) Quantitation of the bioluminescent signal (BLI) expressed as average radiant intensity, obtained using the IVIS spectrum 200 equipment (N = 2 plates/group). Data expressed as mean ± s.e.m. Comparison of emitted BLI between groups: (B: Buffer; F: free oligo‐FdU; T‐F: T22‐GFP‐H6‐FdU). P‐values for statistical differences: T‐F vs. B, **P = 0.001 (green line, panel D); F vs. B, *P = 0.011 (red line); T‐F vs. F at *P = 0.02 (black line, panel D). Mann–Whitney U‐test.

- A, B
Reduction in the number of formed spheroids (white arrows, optical microscope) generated by 1 × 106 disaggregated cells (cultured in stem cell‐conditioned media and low‐adhesion plates) obtained from CXCR4+ M5 subcutaneous tumors, 24 h after 100 μg T22‐GFP‐H6‐FdU intravenous doses, for 2 consecutive days, as compared to Buffer‐treated or free oligo‐FdU‐treated mice (mean ± s.e.m., N = 8; 2 mice/group; 4 plates/mouse). Scale bar, 100 μm. Comparison of spheroid formation between groups: (B: Buffer; F: free oligo‐FdU; T‐F: T22‐GFP‐H6‐FdU). P‐values for statistical differences: T‐F vs. B, **P = 0.001 (green line, Panel B); F vs. B, *P = 0.012 (red line); T‐F vs. F, **P = 0.001 (black line). Mann–Whitney U‐test.
- C, D
Reduction in tumor re‐initiation capacity after subcutaneous inoculation of 5 × 106 cells in NSG mice (N = 4 tumors/group) derived from disaggregated tumor cells obtained from SC tumors 10 days after administration of 100 μg T22‐GFP‐H6‐FdU intravenous doses, for 2 consecutive days, as compared to free oligo‐FdU‐treated or Buffer‐treated mice. Recording of the number and size of positive tumors (black arrows, N = 4; 2 mice/group, 2 injection points/mouse).
- E–G
(E) Representative images of tumor emboli intravasation determined by microscopic analyses of H&E‐stained tumor sections (N = 5/group). (F) T22‐GFP‐H6‐FdU‐induced reduction in the number of intravasated tumor emboli (black arrows) in peri‐tumoral vessels of the M5‐orthotopic primary tumor (E: optical images; F: emboli number quantitation) and reduction in CXCR4 expression in these emboli (G), treated 7 days after tumor cell implantation with 100 μg T22‐GFP‐H6‐FdU intravenous doses, for 2 consecutive days, as compared to Buffer‐treated or free oligo‐FdU‐treated mice. Tumor emboli counting in 10 high‐power field at 200× magnification in H&E‐stained sections from each tumor (mean ± s.e.m., N = 5/group). Comparison of tumor emboli number between groups: (B: Buffer; F: free oligo‐FdU; T‐F: T22‐GFP‐H6‐FdU). P‐values for statistical differences: T‐F vs. B, *P = 0.038 (green line), T‐F vs. F, *P = 0.016 (black line). Scale bar, 100 μm. (G) CXCR4 expression per tumor emboli determined by using anti‐CXCR4 IHC and calculating H‐score (multiplying percent of CXCR4+ cells out of total cell number in the emboli area by their staining intensity, scoring each from 0 to 3 (where 3 is the maximal intensity) per tumor emboli area (mean ± s.e.m., N = 5 mice/group). Comparison of CXCR4 H‐score between groups: (B: Buffer; T‐F: T22‐GFP‐H6‐FdU). P‐values for statistical differences: T‐F vs. B at *P = 0.027 (green line). Mann–Whitney U‐test.

- A
T22‐GFP‐H6‐FdU prevents metastases in the CXCR4+ patient‐derived M5 model by potently reducing the total and mean number of liver, lung, and peritoneal Mets, as recorded in H&E‐stained histology sections at the end of treatment, in comparison with free oligo‐FdU or Buffer treatment. In contrast, the number of LN Mets is not reduced after T22‐GFP‐H6‐FdU or free oligo‐FdU administration (N = 6 mice per Buffer group; N = 7 mice per free oligo‐FdU group; and N = 8 mice per T22‐GFP‐H6‐FdU group; 3 samples/mouse). Data expressed as mean ± s.e.m. Comparison of metastatic foci number by site between groups: (B: Buffer; F: free oligo‐FdU; T‐F: T22‐GFP‐H6‐FdU). P‐values for statistical differences: T‐F vs. B: *P = 0.006 for LN Mets; *P = 0.001 for LV Mets; *P = 0.003 for LG Mets; *P = 0.001 for PTN Mets (green lines), F vs. B: *P = 0.015 for PTN Mets (red line), T‐F vs. F: *P = 0.001 for LV Mets, *P = 0.022 for PTN Mets (black line). Mann–Whitney U‐test. See Table 1 for the recording of the percent of metastasis‐free mice (mice with undetectable metastases at the end of treatment, and therefore with an absence of CXCR4+ tumor cells) after T22‐GFP‐H6‐FdU treatment. Also, Table 1 describes the reduction in mean foci number and foci size in metastasis‐positive mice after T22‐GFP‐H6‐FdU treatment, as compared to Buffer or free oligo‐FdU.
- B
T22‐GFP‐H6‐FdU induces a higher reduction in CXCR4+ cancer cell fraction (CXCR4+ CCF) in liver, lung, and peritoneal metastatic tissue, at the end of treatment, than free oligo‐FdU, as measured by anti‐CXCR4 IHC. In contrast, T22‐GFP‐H6‐FdU or free oligo‐FdU does not reduce the CXCR4+ CCF in LN Mets or primary tumor tissue after therapy (N = 6 mice per Buffer group; N = 7 mice per free oligo‐FdU group; and N = 8 mice per T22‐GFP‐H6‐FdU group; 3 samples/mouse). Data expressed as mean ± s.e.m. Comparison of remaining CXCR4+ CCF by site between groups: (B: Buffer; F: free oligo‐FdU; T‐F: T22‐GFP‐H6‐FdU). P‐values for statistical differences: T‐F vs. B: *P = 0.012 for LV Mets, *P = 0.027 for LG Mets; *P = 0.038 for PTN Mets (green lines), T‐F vs. F: *P = 0.013 for LV Mets (black line). Mann–Whitney U‐test.
- C
Representative CXCR4 IHC images of the reduction in CXCR4+ CCF induced by T22‐GFP‐H6‐FdU (or its absence in free oligo‐FdU mice) at the end of treatment, in the M5 patient‐derived CRC model, which quantitation is reported in panel (B). In the M5 model, the highest reduction in foci number and size occurs in liver metastases, which show the highest reduction in CXCR4+ CCF. Note the correlation between the reduction in CXCR4+ CCF induced by T22‐GFP‐H6‐FdU and its antimetastatic effect at each site, measured as number of liver, lung, or peritoneal Mets (Table 1) in the M5 metastatic CRC models [as it happens in the SW1417 model (Appendix Fig S8)]. Note in both Table 1 and Appendix Table S1 that 83% of mice in the T22‐GFP‐H6‐FdU group remained free of liver, lung, or peritoneal metastases at the end of treatment in the SW1417 CRC model, whereas in the M5 CRC model these parameters were in the 38–63% range. Scale bar, 100 μm. Asterisks, tumor tissue; N, normal tissue; LN, lymphatic metastasis.

- A
Undetectable T22‐GFP‐H6‐FdU emitted fluorescence in normal tissues, except for a transient accumulation 5 h after a 100 μg dose in the liver, which disappears at 24 h. Liver emitted fluorescence is transient and significantly lower than the one registered in tumor tissue. Tumor/Liver ratio = 7.5 (see tumor intensity in Fig 2B, which was registered in the same experiment; N = 5 mice/group). Scale bar, 1 cm. Color key, radiant efficiency units.
- B
Representative images depicting the level of DNA double‐strand break (DSB) induction in histologically normal bone marrow 5 h after treatment, as measured by anti‐γ‐H2AX, which is higher in free oligo‐FdU‐treated mice than in T22‐GFP‐H6‐FdU (P = 0.047). Low level of cells containing DSBs in histologically normal kidney after T22‐GFP‐H6‐FdU or free oligo‐FdU treatment, a finding occurring in all normal tissues analyzed (N = 50, 5 mice/group; 10 fields/mouse). Scale bar, 100 μm.
- C
Representative images showing lack of histopathological alterations in H&E‐stained tissue or apoptotic induction in H&E‐stained samples of CXCR4+ (bone marrow) and CXCR4− (brain, kidney, liver, lung, and heart) normal tissues 24 h after the administration of a 100 μg dose of T22‐GFP‐H6‐FdU or an equimolecular dose of free oligo‐FdU (N = 5/group). Note that the transient nanoconjugate distribution to liver or the DNA damage induced in bone marrow does not lead to cytotoxicity on these non‐tumor tissues (N = 50, 5 mice/group; 10 fields/mouse). Scale bar, 100 μm.
- D
Lack of differences in body weight among groups registered along time in the SW1417‐derived CCR model and the regression of metastases protocol (mean ± s.e.m., N = 10 mice/group).
- E
Lack of differences in body mouse weight among groups registered along time in the SW1417 cell line‐derived model and the prevention of metastasis protocol [mean ± s.e.m., Buffer (N = 11; free oligo‐FdU (N = 12), T22‐GFP‐H6‐FdU (N = 12)].
- F
Lack of differences in body mouse weight among groups registered along time in the M5 patient‐derived model and the prevention of metastasis protocol [mean ± s.e.m., Buffer (N = 6); free oligo‐FdU (N = 17); T22‐GFP‐H6‐FdU (N = 8)].
Similar articles
-
An Auristatin nanoconjugate targeting CXCR4+ leukemic cells blocks acute myeloid leukemia dissemination.J Hematol Oncol. 2020 Apr 15;13(1):36. doi: 10.1186/s13045-020-00863-9. J Hematol Oncol. 2020. PMID: 32295630 Free PMC article.
-
GSDMD-dependent pyroptotic induction by a multivalent CXCR4-targeted nanotoxin blocks colorectal cancer metastases.Drug Deliv. 2022 Dec;29(1):1384-1397. doi: 10.1080/10717544.2022.2069302. Drug Deliv. 2022. PMID: 35532120 Free PMC article.
-
Rational engineering of a human GFP-like protein scaffold for humanized targeted nanomedicines.Acta Biomater. 2021 Aug;130:211-222. doi: 10.1016/j.actbio.2021.06.001. Epub 2021 Jun 9. Acta Biomater. 2021. PMID: 34116228
-
Cancer stem cells as new therapeutic target to prevent tumour progression and metastasis.Front Biosci (Elite Ed). 2010 Jan 1;2(2):602-13. doi: 10.2741/e117. Front Biosci (Elite Ed). 2010. PMID: 20036905 Review.
-
The axis of evil in the fight against cancer.Rom J Intern Med. 2011;49(4):319-25. Rom J Intern Med. 2011. PMID: 22568277 Review.
Cited by
-
Self-assembling protein nanocarrier for selective delivery of cytotoxic polypeptides to CXCR4+ head and neck squamous cell carcinoma tumors.Acta Pharm Sin B. 2022 May;12(5):2578-2591. doi: 10.1016/j.apsb.2021.09.030. Epub 2021 Oct 14. Acta Pharm Sin B. 2022. PMID: 35646535 Free PMC article.
-
Clinical Implications of Colorectal Cancer Stem Cells in the Age of Single-Cell Omics and Targeted Therapies.Gastroenterology. 2021 May;160(6):1947-1960. doi: 10.1053/j.gastro.2020.12.080. Epub 2021 Feb 19. Gastroenterology. 2021. PMID: 33617889 Free PMC article. Review.
-
An In Silico Methodology That Facilitates Decision Making in the Engineering of Nanoscale Protein Materials.Int J Mol Sci. 2022 Apr 29;23(9):4958. doi: 10.3390/ijms23094958. Int J Mol Sci. 2022. PMID: 35563346 Free PMC article.
-
Structural Stabilization of Clinically Oriented Oligomeric Proteins During their Transit through Synthetic Secretory Amyloids.Adv Sci (Weinh). 2024 Jun;11(21):e2309427. doi: 10.1002/advs.202309427. Epub 2024 Mar 19. Adv Sci (Weinh). 2024. PMID: 38501900 Free PMC article.
-
Nano-Based Approved Pharmaceuticals for Cancer Treatment: Present and Future Challenges.Biomolecules. 2022 Jun 4;12(6):784. doi: 10.3390/biom12060784. Biomolecules. 2022. PMID: 35740909 Free PMC article. Review.
References
-
- Balkwill F (2004) The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol 14: 171–179 - PubMed
-
- Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T (2005) Migrating cancer stem cells‐an integrated concept of malignant tumour progression. Nat Rev Cancer 5: 744–749 - PubMed
-
- Céspedes MV, Espina C, García‐Cabezas MA, Trias M, Boluda A, Gómez del Pulgar MT, Sancho FJ, Nistal M, Lacal JC, Mangues R (2007) Orthotopic microinjection of human colon cancer cells in nude mice induces tumor foci in all clinically relevant metastatic sites. Am J Pathol 170: 1077–1085 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

