Utilization of rare codon-rich markers for screening amino acid overproducers
- PMID: 30190534
- PMCID: PMC6127279
- DOI: 10.1038/s41467-018-05830-0
Utilization of rare codon-rich markers for screening amino acid overproducers
Abstract
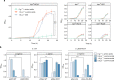
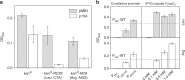
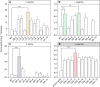
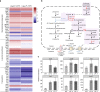
The translation of rare codons relies on their corresponding rare tRNAs, which could not be fully charged under amino acid starvation. Theoretically, disrupted or retarded translation caused by the lack of charged rare tRNAs can be partially restored by feeding or intracellular synthesis of the corresponding amino acids. Inspired by this assumption, we develop a screening or selection system for obtaining overproducers of a target amino acid by replacing its common codons with the corresponding synonymous rare alternative in the coding sequence of selected reporter proteins or antibiotic-resistant markers. Results show that integration of rare codons can inhibit gene translations in a frequency-dependent manner. As a proof-of-concept, Escherichia coli strains overproducing L-leucine, L-arginine or L-serine are successfully selected from random mutation libraries. The system is also applied to Corynebacterium glutamicum to screen out L-arginine overproducers. This strategy sheds new light on obtaining and understanding amino acid overproduction strains.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Tonouchi, N. & Ito, H. Present global situation of amino acids in industry. In Amino Acid Fermentation (eds Yokota, A. & Ikeda, M.) (Springer, New York, 2017). - PubMed
-
- Tatsumi, N. & Inui, M. Corynebacterium Glutamicum: Biology and Biotechnology (Springer, New York, 2012).
-
- Shive, W. & Skinner, C. G. Amino acid analogues. In Metabolic Inhibitors: A Comprehensive Treatise (eds Hochster, R. & Quastel, J.) (Academic Press, Cambridge, MA, 2012).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

