Tracking of Host Defenses and Phylogeny During the Radiation of Neotropical Inga-Feeding Sawflies (Hymenoptera; Argidae)
- PMID: 30190723
- PMCID: PMC6116116
- DOI: 10.3389/fpls.2018.01237
Tracking of Host Defenses and Phylogeny During the Radiation of Neotropical Inga-Feeding Sawflies (Hymenoptera; Argidae)
Abstract
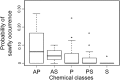
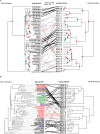
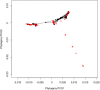
Coevolutionary theory has long predicted that the arms race between plants and herbivores is a major driver of host selection and diversification. At a local scale, plant defenses contribute significantly to the structure of herbivore assemblages and the high alpha diversity of plants in tropical rain forests. However, the general importance of plant defenses in host associations and divergence at regional scales remains unclear. Here, we examine the role of plant defensive traits and phylogeny in the evolution of host range and species divergence in leaf-feeding sawflies of the family Argidae associated with Neotropical trees in the genus Inga throughout the Amazon, the Guiana Shield and Panama. Our analyses show that the phylogenies of both the sawfly herbivores and their Inga hosts are congruent, and that sawflies radiated at approximately the same time, or more recently than their Inga hosts. Analyses controlling for phylogenetic effects show that the evolution of host use in the sawflies associated with Inga is better correlated with Inga chemistry than with Inga phylogeny, suggesting a pattern of delayed host tracking closely tied to host chemistry. Finally, phylogenetic analyses show that sister species of Inga-sawflies are dispersed across the Neotropics, suggesting a role for allopatric divergence and vicariance in Inga diversification. These results are consistent with the idea that host defensive traits play a key role not only in structuring the herbivore assemblages at a single site, but also in the processes shaping host association and species divergence at a regional scale.
Keywords: Inga; coevolution; defense traits; herbivores; host tracking; plant–insect interactions; sawflies; tropical rain forests.
Figures

Similar articles
-
Coevolutionary arms race versus host defense chase in a tropical herbivore-plant system.Proc Natl Acad Sci U S A. 2017 Sep 5;114(36):E7499-E7505. doi: 10.1073/pnas.1707727114. Epub 2017 Aug 21. Proc Natl Acad Sci U S A. 2017. PMID: 28827317 Free PMC article.
-
Consequences of interspecific variation in defenses and herbivore host choice for the ecology and evolution of Inga, a speciose rainforest tree.Oecologia. 2018 Jun;187(2):361-376. doi: 10.1007/s00442-018-4080-z. Epub 2018 Feb 10. Oecologia. 2018. PMID: 29428967 Review.
-
The evolution of antiherbivore defenses and their contribution to species coexistence in the tropical tree genus Inga.Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18073-8. doi: 10.1073/pnas.0904786106. Epub 2009 Sep 21. Proc Natl Acad Sci U S A. 2009. PMID: 19805183 Free PMC article.
-
Quantitative and qualitative shifts in defensive metabolites define chemical defense investment during leaf development in Inga, a genus of tropical trees.Ecol Evol. 2016 Jan 8;6(2):478-92. doi: 10.1002/ece3.1896. eCollection 2016 Jan. Ecol Evol. 2016. PMID: 26843932 Free PMC article.
-
Host specificity of insect herbivores in tropical forests.Proc Biol Sci. 2005 Jun 7;272(1568):1083-90. doi: 10.1098/rspb.2004.3023. Proc Biol Sci. 2005. PMID: 16024368 Free PMC article. Review.
Cited by
-
phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things).PeerJ. 2024 Jan 5;12:e16505. doi: 10.7717/peerj.16505. eCollection 2024. PeerJ. 2024. PMID: 38192598 Free PMC article.
-
Great chemistry between us: The link between plant chemical defenses and butterfly evolution.Ecol Evol. 2021 May 27;11(13):8595-8613. doi: 10.1002/ece3.7673. eCollection 2021 Jul. Ecol Evol. 2021. PMID: 34257918 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

