A resting state fMRI analysis pipeline for pooling inference across diverse cohorts: an ENIGMA rs-fMRI protocol
- PMID: 30191514
- PMCID: PMC6401353
- DOI: 10.1007/s11682-018-9941-x
A resting state fMRI analysis pipeline for pooling inference across diverse cohorts: an ENIGMA rs-fMRI protocol
Abstract
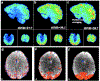
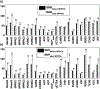
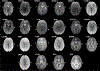
Large-scale consortium efforts such as Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) and other collaborative efforts show that combining statistical data from multiple independent studies can boost statistical power and achieve more accurate estimates of effect sizes, contributing to more reliable and reproducible research. A meta- analysis would pool effects from studies conducted in a similar manner, yet to date, no such harmonized protocol exists for resting state fMRI (rsfMRI) data. Here, we propose an initial pipeline for multi-site rsfMRI analysis to allow research groups around the world to analyze scans in a harmonized way, and to perform coordinated statistical tests. The challenge lies in the fact that resting state fMRI measurements collected by researchers over the last decade vary widely, with variable quality and differing spatial or temporal signal-to-noise ratio (tSNR). An effective harmonization must provide optimal measures for all quality data. Here we used rsfMRI data from twenty-two independent studies with approximately fifty corresponding T1-weighted and rsfMRI datasets each, to (A) review and aggregate the state of existing rsfMRI data, (B) demonstrate utility of principal component analysis (PCA)-based denoising and (C) develop a deformable ENIGMA EPI template based on the representative anatomy that incorporates spatial distortion patterns from various protocols and populations.
Keywords: ENIGMA EPI template; Large multi-site studies; Processing pipelines.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Grants and funding
- U54EB020403, U01MH108148, 2R01EB015611, R01MH112180, R01DA027680, R01MH085646/GF/NIH HHS/United States
- R01 MH116948/MH/NIMH NIH HHS/United States
- R01 EB015611/EB/NIBIB NIH HHS/United States
- FOR2017:KO 4291/4-1 and KR 3822/5-1/German Research Foundation (DFG)
- R01 DA027680/DA/NIDA NIH HHS/United States
- 12S1615N/Postdoctoral Fellow of the Research Foundation - Flanders (FWO)
- R01 MH085646/MH/NIMH NIH HHS/United States
- P41 EB017183/EB/NIBIB NIH HHS/United States
- U01 MH108148/MH/NIMH NIH HHS/United States
- U54 EB020403/EB/NIBIB NIH HHS/United States
- R01 MH112180/MH/NIMH NIH HHS/United States
- R01 NS088040/NS/NINDS NIH HHS/United States
- FOR2107: DA 1151/5-1/German Research Foundation
- FOR2107:KI 588/15-1/German Research Foundation
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

