The metagenome of the female upper reproductive tract
- PMID: 30192933
- PMCID: PMC6177736
- DOI: 10.1093/gigascience/giy107
The metagenome of the female upper reproductive tract
Abstract
Background: The human uterus is traditionally believed to be sterile, while the vaginal microbiota plays an important role in fending off pathogens. Emerging evidence demonstrates the presence of bacteria beyond the vagina. However, a microbiome-wide metagenomic analysis characterizing the diverse microbial communities has been lacking.
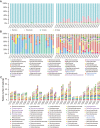
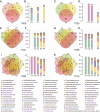
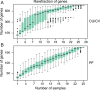
Results: We performed shotgun-sequencing of 52 samples from the cervical canal and the peritoneal fluid of Chinese women of reproductive age using the Illumina platform. Direct annotation of sequencing reads identified the taxonomy of bacteria, archaea, fungi and viruses, confirming and extending the results from our previous study. We replicated our previous findings in another 24 samples from the vagina, the cervical canal, the uterus and the peritoneal fluid using the BGISEQ-500 platform revealing that microorganisms in the samples from the same individuals were largely shared in the entire reproductive tract. Human sequences made up more than 99% of the 20GB raw data. After filtering, vaginal microorganisms were well covered in the generated reproductive tract gene catalogue, while the more diverse upper reproductive tract microbiota would require greater depth of sequencing and more samples to meet the full coverage scale.
Conclusions: We provide novel detailed data on the microbial composition of a largely unchartered body site, the female reproductive tract. Our results indicated the presence of an intra-individual continuum of microorganisms that gradually changed from the vagina to the peritoneal fluid. This study also provides a framework for understanding the implications of the composition and functional potential of the distinct microbial ecosystems of the female reproductive tract in relation to health and disease.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

