Single molecule tracking reveals that the bacterial SMC complex moves slowly relative to the diffusion of the chromosome
- PMID: 30192981
- PMCID: PMC6125636
- DOI: 10.1093/nar/gky581
Single molecule tracking reveals that the bacterial SMC complex moves slowly relative to the diffusion of the chromosome
Abstract
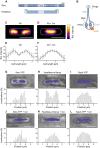
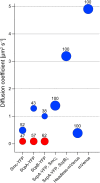
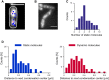
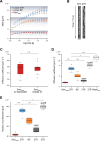
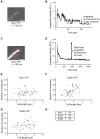
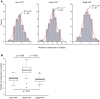
Structural Maintenance of Chromosomes (SMC) proteins and their complex partners (ScpA and ScpB in many bacteria) are involved in chromosome compaction and segregation in all kinds of organisms. We employed single molecule tracking (SMT), tracking of chromosomal loci, and single molecule counting in Bacillus subtilis to show that in slow growing cells, ∼30 Smc dimers move throughout the chromosome in a constrained mode, while ∼60 ScpA and ScpB molecules travel together in a complex, but independently of the nucleoid. Even an Smc truncation that lacks the ATP binding head domains still scans the chromosome, highlighting the importance of coiled coil arm domains. When forming a complex, 10-15 Smc/ScpAB complexes become essentially immobile, moving slower than chromosomal loci. Contrarily, SMC-like protein RecN, which forms assemblies at DNA double strand breaks, moves faster than chromosome sites. In the absence of Smc, chromosome sites investigated were less mobile than in wild type cells, indicating that Smc contributes to chromosome dynamics. Thus, our data show that Smc/ScpAB clusters occur at several sites on the chromosome and contribute to chromosome movement.
Figures
References
-
- Uhlmann F. SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 2016; 17:399–412. - PubMed
-
- Hirano T. Condensin-based chromosome organization from bacteria to vertebrates. Cell. 2016; 164:847–857. - PubMed
-
- Graumann P.L. Chromosome architecture and segregation in prokaryotic cells. J. Mol. Microbiol. Biotechnol. 2014; 24:291–300. - PubMed
-
- Löwe J., Cordell S.C., van den Ent F.. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol. 2001; 306:25–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

