Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-κB Signaling after Nuclear DNA Damage
- PMID: 30193098
- PMCID: PMC6127031
- DOI: 10.1016/j.molcel.2018.07.034
Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-κB Signaling after Nuclear DNA Damage
Abstract
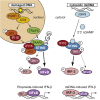
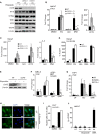
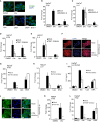
DNA damage can be sensed as a danger-associated molecular pattern by the innate immune system. Here we find that keratinocytes and other human cells mount an innate immune response within hours of etoposide-induced DNA damage, which involves the DNA sensing adaptor STING but is independent of the cytosolic DNA receptor cGAS. This non-canonical activation of STING is mediated by the DNA binding protein IFI16, together with the DNA damage response factors ATM and PARP-1, resulting in the assembly of an alternative STING signaling complex that includes the tumor suppressor p53 and the E3 ubiquitin ligase TRAF6. TRAF6 catalyzes the formation of K63-linked ubiquitin chains on STING, leading to the activation of the transcription factor NF-κB and the induction of an alternative STING-dependent gene expression program. We propose that STING acts as a signaling hub that coordinates a transcriptional response depending on its mode of activation.
Keywords: DNA damage; IFI16; STING; TRAF6; etoposide; innate immunity; interferon; p53; ubiquitin.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Aglipay J.A., Lee S.W., Okada S., Fujiuchi N., Ohtsuka T., Kwak J.C., Wang Y., Johnstone R.W., Deng C., Qin J., Ouchi T. A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene. 2003;22:8931–8938. - PubMed
-
- Banin S., Moyal L., Shieh S., Taya Y., Anderson C.W., Chessa L., Smorodinsky N.I., Prives C., Reiss Y., Shiloh Y., Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

