Matrix mineralization controls gene expression in osteoblastic cells
- PMID: 30193837
- PMCID: PMC6185740
- DOI: 10.1016/j.yexcr.2018.09.005
Matrix mineralization controls gene expression in osteoblastic cells
Abstract
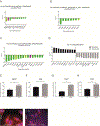
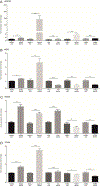
Osteoblasts are adherent cells, and under physiological conditions they attach to both mineralized and non-mineralized osseous surfaces. However, how exactly osteoblasts respond to these different osseous surfaces is largely unknown. Our hypothesis was that the state of matrix mineralization provides a functional signal to osteoblasts. To assess the osteoblast response to mineralized compared to demineralized osseous surfaces, we developed and validated a novel tissue surface model. We demonstrated that with the exception of the absence of mineral, the mineralized and demineralized surfaces were similar in molecular composition as determined, for example, by collagen content and maturity. Subsequently, we used the human osteoblastic cell line MG63 in combination with genome-wide gene set enrichment analysis (GSEA) to record and compare the gene expression signatures on mineralized and demineralized surfaces. Assessment of the 5 most significant gene sets showed on mineralized surfaces an enrichment exclusively of genes sets linked to protein synthesis, while on the demineralized surfaces 3 of the 5 enriched gene sets were associated with the matrix. Focusing on these three gene sets, we observed not only the expected structural components of the bone matrix, but also gene products, such as HMCN1 or NID2, that are likely to act as temporal migration guides. Together, these findings suggest that in osteoblasts mineralized and demineralized osseous surfaces favor intracellular protein production and matrix formation, respectively. Further, they demonstrate that the mineralization state of bone independently controls gene expression in osteoblastic cells.
Keywords: Bone; Bone matrix; Bone mineral; Gene Regulation; Osteoblast.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
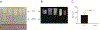



References
-
- Long F, Building strong bones: molecular regulation of the osteoblast lineage, Nat Rev Mol Cell Biol 13 (2011) 27–38. - PubMed
-
- Safadi FF, Barbe MF, Abdelmagid SM, Rico MC, Aswad RA, Litvin J, Popoff SN, Bone Structure, Development and Bone Biology, in: Khurana JS (Ed.), Bone Pathology, Humana Press, 2009.
-
- Boyan BD, Schwartz Z, Boskey AL, The importance of mineral in bone and mineral research, Bone 27 (2000) 341–342. - PubMed
-
- Boskey AL, Biomineralization: an overview, Connect Tissue Res 44 Suppl 1 (2003) 5–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

