Antibodies Set Boundaries Limiting Microbial Metabolite Penetration and the Resultant Mammalian Host Response
- PMID: 30193848
- PMCID: PMC6162337
- DOI: 10.1016/j.immuni.2018.08.004
Antibodies Set Boundaries Limiting Microbial Metabolite Penetration and the Resultant Mammalian Host Response
Abstract
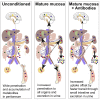
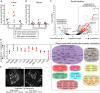
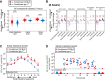
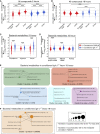
Although the mammalian microbiota is well contained within the intestine, it profoundly shapes development and metabolism of almost every host organ. We questioned the range and depth of microbial metabolite penetration into the host, and how this is modulated by intestinal immunity. Chemically identical microbial and host metabolites were distinguished by stable isotope tracing from 13C-labeled live non-replicating Escherichia coli, differentiating 12C host isotopes with high-resolution mass spectrometry. Hundreds of endogenous microbial compounds penetrated 23 host tissues and fluids after intestinal exposure: subsequent 12C host metabolome signatures included lipidemia, reduced glycolysis, and inflammation. Penetrant bacterial metabolites from the small intestine were rapidly cleared into the urine, whereas induced antibodies curtailed microbial metabolite exposure by accelerating intestinal bacterial transit into the colon where metabolite transport mechanisms are limiting. Pervasive penetration of microbial molecules can cause extensive host tissue responses: these are limited by immune and non-immune intestinal mucosal adaptations to the microbiota.
Keywords: (13)C-isotope tracing; IgA; colonization; germ free; inflammation; intestinal transit; lipidemia; metabolomics; microbial metabolites; microbiota.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
Gone with the Antibody.Immunity. 2018 Sep 18;49(3):386-388. doi: 10.1016/j.immuni.2018.09.003. Immunity. 2018. PMID: 30231978
References
-
- Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. - PubMed
-
- Beatty D.W., Napier B., Sinclair-Smith C.C., McCabe K., Hughes E.J. Secretory IgA synthesis in Kwashiorkor. J. Clin. Lab. Immunol. 1983;12:31–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

