Mutually inhibitory Ras-PI(3,4)P2 feedback loops mediate cell migration
- PMID: 30194235
- PMCID: PMC6166812
- DOI: 10.1073/pnas.1809039115
Mutually inhibitory Ras-PI(3,4)P2 feedback loops mediate cell migration
Erratum in
-
Correction for Li et al., Mutually inhibitory Ras-PI(3,4)P2 feedback loops mediate cell migration.Proc Natl Acad Sci U S A. 2018 Oct 23;115(43):E10286. doi: 10.1073/pnas.1816155115. Epub 2018 Oct 15. Proc Natl Acad Sci U S A. 2018. PMID: 30322942 Free PMC article. No abstract available.
Abstract
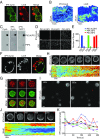
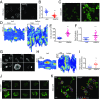
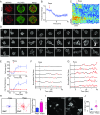
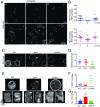
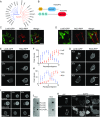
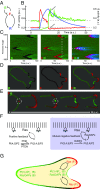
Signal transduction and cytoskeleton networks in a wide variety of cells display excitability, but the mechanisms are poorly understood. Here, we show that during random migration and in response to chemoattractants, cells maintain complementary spatial and temporal distributions of Ras activity and phosphatidylinositol (3,4)-bisphosphate [PI(3,4)P2]. In addition, depletion of PI(3,4)P2 by disruption of the 5-phosphatase, Dd5P4, or by recruitment of 4-phosphatase INPP4B to the plasma membrane, leads to elevated Ras activity, cell spreading, and altered migratory behavior. Furthermore, RasGAP2 and RapGAP3 bind to PI(3,4)P2, and the phenotypes of cells lacking these genes mimic those with low PI(3,4)P2 levels, providing a molecular mechanism. These findings suggest that Ras activity drives PI(3,4)P2 down, causing the PI(3,4)P2-binding GAPs to dissociate from the membrane, further activating Ras, completing a positive-feedback loop essential for excitability. Consistently, a computational model incorporating such a feedback loop in an excitable network model accurately simulates the dynamic distributions of active Ras and PI(3,4)P2 as well as cell migratory behavior. The mutually inhibitory Ras-PI(3,4)P2 mechanisms we uncovered here provide a framework for Ras regulation that may play a key role in many physiological processes.
Keywords: chemotaxis; excitability; phosphoinositides; positive feedback loop; signal transduction.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Weiger MC, Parent CA. Phosphoinositides in chemotaxis. Subcell Biochem. 2012;59:217–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

