Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3'-overhang
- PMID: 30194297
- PMCID: PMC6128929
- DOI: 10.1038/s41467-018-06129-w
Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3'-overhang
Abstract
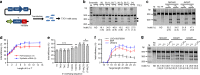
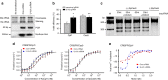
Genome editing has been harnessed through the development of CRISPR system, and the CRISPR from Prevotella and Francisella 1 (Cpf1) system has emerged as a promising alternative to CRISPR-Cas9 for use in various circumstances. Despite the inherent multiple advantages of Cpf1 over Cas9, the adoption of Cpf1 has been unsatisfactory because of target-dependent insufficient indel efficiencies. Here, we report an engineered CRISPR RNA (crRNA) for highly efficient genome editing by Cpf1, which includes a 20-base target-complementary sequence and a uridinylate-rich 3'-overhang. When the crRNA is transcriptionally produced, crRNA with a 20-base target-complementary sequence plus a U4AU4 3'-overhang is the optimal configuration. U-rich crRNA also maximizes the utility of the AsCpf1 mutants and multiplexing genome editing using mRNA as the source of multiple crRNAs. Furthermore, U-rich crRNA enables a highly safe and specific genome editing using Cpf1 in human cells, contributing to the enhancement of a genome-editing toolbox.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

