Cell type-resolved human lung lipidome reveals cellular cooperation in lung function
- PMID: 30194354
- PMCID: PMC6128932
- DOI: 10.1038/s41598-018-31640-x
Cell type-resolved human lung lipidome reveals cellular cooperation in lung function
Abstract
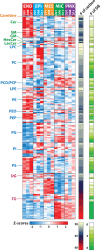
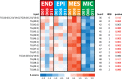
Cell type-resolved proteome analyses of the brain, heart and liver have been reported, however a similar effort on the lipidome is currently lacking. Here we applied liquid chromatography-tandem mass spectrometry to characterize the lipidome of major lung cell types isolated from human donors, representing the first lipidome map of any organ. We coupled this with cell type-resolved proteomics of the same samples (available at Lungmap.net). Complementary proteomics analyses substantiated the functional identity of the isolated cells. Lipidomics analyses showed significant variations in the lipidome across major human lung cell types, with differences most evident at the subclass and intra-subclass (i.e. total carbon length of the fatty acid chains) level. Further, lipidomic signatures revealed an overarching posture of high cellular cooperation within the human lung to support critical functions. Our complementary cell type-resolved lipid and protein datasets serve as a rich resource for analyses of human lung function.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

