Neuronal microRNA regulation in Experimental Autoimmune Encephalomyelitis
- PMID: 30194392
- PMCID: PMC6128870
- DOI: 10.1038/s41598-018-31542-y
Neuronal microRNA regulation in Experimental Autoimmune Encephalomyelitis
Abstract
Multiple sclerosis (MS) is an autoimmune, neurodegenerative disease but the molecular mechanisms underlying neurodegenerative aspects of the disease are poorly understood. microRNAs (miRNAs) are powerful regulators of gene expression that regulate numerous mRNAs simultaneously and can thus regulate programs of gene expression. Here, we describe miRNA expression in neurons captured from mice subjected to experimental autoimmune encephalomyelitis (EAE), a model of central nervous system (CNS) inflammation. Lumbar motor neurons and retinal neurons were laser captured from EAE mice and miRNA expression was assessed by next-generation sequencing and validated by qPCR. We describe 14 miRNAs that are differentially regulated in both neuronal subtypes and determine putative mRNA targets though in silico analysis. Several upregulated neuronal miRNAs are predicted to target pathways that could mediate repair and regeneration during EAE. This work identifies miRNAs that are affected by inflammation and suggests novel candidates that may be targeted to improve neuroprotection in the context of pathological inflammation.
Conflict of interest statement
Dr. Bar-Or has participated as a speaker in meetings sponsored by and received consulting fees and/or grant support from: Atara Biotherapeutics, Biogen Idec, Celgene/Receptos, Genentech/Roche, GlaxoSmithKline, MAPI, Medimmune, Merck/EMD Serono, Novartis, Sanofi-Genzyme.
Figures
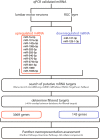
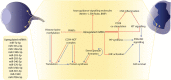
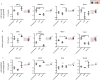
References
-
- Dutta, R. & Trapp, B. D. Pathogenesis of axonal and neuronal damage in multiple sclerosis. Neurology 68, S22–31; discussion S43–54 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

