Differential Co-expression and Regulatory Network Analysis Uncover the Relapse Factor and Mechanism of T Cell Acute Leukemia
- PMID: 30195757
- PMCID: PMC6023839
- DOI: 10.1016/j.omtn.2018.05.003
Differential Co-expression and Regulatory Network Analysis Uncover the Relapse Factor and Mechanism of T Cell Acute Leukemia
Abstract
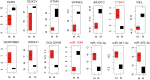
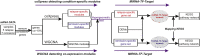
The pediatric T cell acute lymphoblastic leukemia (T-ALL) still remains a cancer with worst prognosis for high recurrence. Massive studies were conducted for the leukemia relapse based on diagnosis and relapse paired samples. However, the initially diagnostic samples may contain the relapse information and mechanism, which were rarely studied. In this study, we collected mRNA and microRNA (miRNA) data from initially diagnosed pediatric T-ALL samples with their relapse or remission status after treatment. Integrated differential co-expression and miRNA-transcription factor (TF)-gene regulatory network analyses were used to reveal the possible relapse mechanisms for pediatric T-ALL. We detected miR-1246/1248 and NOTCH2 served as key nodes in the relapse network, and they combined with TF WT1/SOX4/REL to form regulatory modules that influence the progress of T-ALL. A regulatory loop miR-429-MYCN-MFHAS1 was found potentially associated with the remission of T-ALL. Furthermore, we proved miR-1246/1248 combined with NOTCH2 could promote cell proliferation in the T-ALL cell line by experiments. Meanwhile, analysis based on the miRNA-drug relationships demonstrated that drugs 5-fluorouracil, ascorbate, and trastuzumab targeting miR-1246 could serve as potential supplements for the standard therapy. In conclusion, our findings revealed the potential molecular mechanisms of T-ALL relapse by the combination of co-expression and regulatory network, and they provide preliminary clues for precise treatment of T-ALL patients.
Keywords: T-ALL; co-expression; drug; network; relapse.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Simultaneous changes in expression levels of BAALC and miR-326: a novel prognostic biomarker for childhood ALL.Jpn J Clin Oncol. 2020 Jun 10;50(6):671-678. doi: 10.1093/jjco/hyaa025. Jpn J Clin Oncol. 2020. PMID: 32129446
-
MicroRNA-106b~25 cluster is upregulated in relapsed MLL-rearranged pediatric acute myeloid leukemia.Oncotarget. 2016 Jul 26;7(30):48412-48422. doi: 10.18632/oncotarget.10270. Oncotarget. 2016. PMID: 27351222 Free PMC article.
-
Integrating microRNA and mRNA expression in rapamycin-treated T-cell acute lymphoblastic leukemia.Pathol Res Pract. 2019 Aug;215(8):152494. doi: 10.1016/j.prp.2019.152494. Epub 2019 Jun 15. Pathol Res Pract. 2019. PMID: 31229277
-
Integrated analysis of microRNA and transcription factors in the bone marrow of patients with acute monocytic leukemia.Oncol Lett. 2021 Jan;21(1):50. doi: 10.3892/ol.2020.12311. Epub 2020 Nov 18. Oncol Lett. 2021. PMID: 33281961 Free PMC article.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
Distance correlation application to gene co-expression network analysis.BMC Bioinformatics. 2022 Feb 21;23(1):81. doi: 10.1186/s12859-022-04609-x. BMC Bioinformatics. 2022. PMID: 35193539 Free PMC article.
-
The role of oncogenic Notch2 signaling in cancer: a novel therapeutic target.Am J Cancer Res. 2019 May 1;9(5):837-854. eCollection 2019. Am J Cancer Res. 2019. PMID: 31218097 Free PMC article. Review.
-
microRNA-1246-containing extracellular vesicles from acute myeloid leukemia cells promote the survival of leukemia stem cells via the LRIG1-meditated STAT3 pathway.Aging (Albany NY). 2021 Apr 23;13(10):13644-13662. doi: 10.18632/aging.202893. Epub 2021 Apr 23. Aging (Albany NY). 2021. PMID: 33893245 Free PMC article.
-
lncRNAs-mRNAs Co-Expression Network Underlying Childhood B-Cell Acute Lymphoblastic Leukaemia: A Pilot Study.Cancers (Basel). 2020 Sep 2;12(9):2489. doi: 10.3390/cancers12092489. Cancers (Basel). 2020. PMID: 32887470 Free PMC article.
-
Identification of potential pathways and microRNA-mRNA networks associated with benzene metabolite hydroquinone-induced hematotoxicity in human leukemia K562 cells.BMC Pharmacol Toxicol. 2022 Apr 2;23(1):20. doi: 10.1186/s40360-022-00556-8. BMC Pharmacol Toxicol. 2022. PMID: 35366954 Free PMC article.
References
-
- Pui C.-H., Robison L.L., Look A.T. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. - PubMed
-
- Aifantis I., Raetz E., Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat. Rev. Immunol. 2008;8:380–390. - PubMed
-
- Seibel N.L., Steinherz P.G., Sather H.N., Nachman J.B., Delaat C., Ettinger L.J., Freyer D.R., Mattano L.A., Jr., Hastings C.A., Rubin C.M. Early postinduction intensification therapy improves survival for children and adolescents with high-risk acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2008;111:2548–2555. - PMC - PubMed
-
- Durinck K., Goossens S., Peirs S., Wallaert A., Van Loocke W., Matthijssens F., Pieters T., Milani G., Lammens T., Rondou P. Novel biological insights in T-cell acute lymphoblastic leukemia. Exp. Hematol. 2015;43:625–639. - PubMed
-
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., 4th, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

