Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification
- PMID: 30195789
- PMCID: PMC6076213
- DOI: 10.1016/j.omtn.2018.06.010
Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification
Abstract
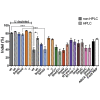
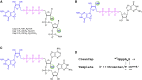
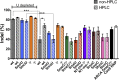
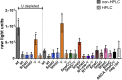
The Cas9/guide RNA (Cas9/gRNA) system is commonly used for genome editing. mRNA expressing Cas9 can induce innate immune responses, reducing Cas9 expression. First-generation Cas9 mRNAs were modified with pseudouridine and 5-methylcytosine to reduce innate immune responses. We combined four approaches to produce more active, less immunogenic second-generation Cas9 mRNAs. First, we developed a novel co-transcriptional capping method yielding natural Cap 1. Second, we screened modified nucleotides in Cas9 mRNA to identify novel modifications that increase Cas9 activity. Third, we depleted the mRNA of uridines to improve mRNA activity. Lastly, we tested high-performance liquid chromatography (HPLC) purification to remove double-stranded RNAs. The activity of these mRNAs was tested in cell lines and primary human CD34+ cells. Cytokines were measured in whole blood and mice. These approaches yielded more active and less immunogenic mRNA. Uridine depletion (UD) most impacted insertion or deletion (indel) activity. Specifically, 5-methoxyuridine UD induced indel frequencies as high as 88% (average ± SD = 79% ± 11%) and elicited minimal immune responses without needing HPLC purification. Our work suggests that uridine-depleted Cas9 mRNA modified with 5-methoxyuridine (without HPLC purification) or pseudouridine may be optimal for the broad use of Cas9 both in vitro and in vivo.
Keywords: ARCA; CRISPR; Cap 1; Cas9; CleanCap; capping; innate immunity; mRNA; uridine depletion.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

