Zinc-Finger Nucleases Induced by HIV-1 Tat Excise HIV-1 from the Host Genome in Infected and Latently Infected Cells
- PMID: 30195798
- PMCID: PMC6023959
- DOI: 10.1016/j.omtn.2018.04.014
Zinc-Finger Nucleases Induced by HIV-1 Tat Excise HIV-1 from the Host Genome in Infected and Latently Infected Cells
Abstract
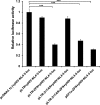
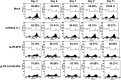
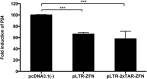
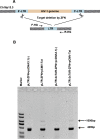
Highly active anti-retroviral therapy (HAART) cannot clear infected cells harboring HIV-1 proviral DNA from HIV-1-infected patients. We previously demonstrated that zinc-finger nucleases (ZFNs) can specifically and efficiently excise HIV-1 proviral DNA from latently infected human T cells by targeting long terminal repeats (LTRs), a novel and alternative antiretroviral strategy for eradicating HIV-1 infection. To prevent unwanted off-target effects from constantly expressed ZFNs, in this study, we engineered the expression of ZFNs under the control of HIV-1 LTR, by which ZFN expression can be activated by the HIV-1 (Trans-Activator of Transcription) Tat protein. Our results show that functional expression of ZFNs induced by Tat excise the integrated proviral DNA of HIV-NL4-3-eGFP in approximately 30% of the population of HIV-1-infected cells. The results from HIV-1-infected human primary T cells and latently infected T cells treated with the inducible ZFNs further validated that proviral DNA can be excised. Taken together, positively regulated expression of ZFNs in the presence of HIV-1 Tat may provide a safer and novel implementation of genome-editing technology for eradicating HIV-1 proviral DNA from infected host cells.
Keywords: HIV-1; Tat; gene therapy; genome editing; genomic excision; inducible; zinc-finger nuclease.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells.Nucleic Acids Res. 2013 Sep;41(16):7771-82. doi: 10.1093/nar/gkt571. Epub 2013 Jun 26. Nucleic Acids Res. 2013. PMID: 23804764 Free PMC article.
-
Genome editing strategies: potential tools for eradicating HIV-1/AIDS.J Neurovirol. 2015 Jun;21(3):310-21. doi: 10.1007/s13365-014-0308-9. Epub 2015 Feb 26. J Neurovirol. 2015. PMID: 25716921 Free PMC article. Review.
-
Gene Therapy with CRISPR/Cas9 Coming to Age for HIV Cure.AIDS Rev. 2017 Oct-Dec;19(3):167-172. AIDS Rev. 2017. PMID: 29019352
-
Zinc finger nuclease: a new approach for excising HIV-1 proviral DNA from infected human T cells.Mol Biol Rep. 2014 Sep;41(9):5819-27. doi: 10.1007/s11033-014-3456-3. Epub 2014 Jun 29. Mol Biol Rep. 2014. PMID: 24973878
-
Origins of Programmable Nucleases for Genome Engineering.J Mol Biol. 2016 Feb 27;428(5 Pt B):963-89. doi: 10.1016/j.jmb.2015.10.014. Epub 2015 Oct 23. J Mol Biol. 2016. PMID: 26506267 Free PMC article. Review.
Cited by
-
Reduce and Control: A Combinatorial Strategy for Achieving Sustained HIV Remissions in the Absence of Antiretroviral Therapy.Viruses. 2020 Feb 8;12(2):188. doi: 10.3390/v12020188. Viruses. 2020. PMID: 32046251 Free PMC article. Review.
-
Knowledge From London and Berlin: Finding Threads to a Functional HIV Cure.Front Immunol. 2021 May 27;12:688747. doi: 10.3389/fimmu.2021.688747. eCollection 2021. Front Immunol. 2021. PMID: 34122453 Free PMC article. Review.
-
Improvements of nuclease and nickase gene modification techniques for the treatment of genetic diseases.Front Genome Ed. 2022 Jul 26;4:892769. doi: 10.3389/fgeed.2022.892769. eCollection 2022. Front Genome Ed. 2022. PMID: 35958050 Free PMC article. Review.
-
Block and Lock HIV Cure Strategies to Control the Latent Reservoir.Front Cell Infect Microbiol. 2020 Aug 14;10:424. doi: 10.3389/fcimb.2020.00424. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32923412 Free PMC article. Review.
-
Grand Challenges on HIV/AIDS in China - The 5th Symposium, Yunnan 2024.Emerg Microbes Infect. 2025 Dec;14(1):2492208. doi: 10.1080/22221751.2025.2492208. Epub 2025 Apr 22. Emerg Microbes Infect. 2025. PMID: 40202047 Free PMC article.
References
-
- Liu H., Ma Y., Su Y., Smith M.K., Liu Y., Jin Y., Gu H., Wu J., Zhu L., Wang N. Emerging trends of HIV drug resistance in Chinese HIV-infected patients receiving first-line highly active antiretroviral therapy: a systematic review and meta-analysis. Clin. Infect. Dis. 2014;59:1495–1502. - PMC - PubMed
-
- Trono D., Van Lint C., Rouzioux C., Verdin E., Barré-Sinoussi F., Chun T.W., Chomont N. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329:174–180. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

