Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm
- PMID: 30197295
- PMCID: PMC6151148
- DOI: 10.1016/j.molcel.2018.08.011
Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm
Abstract
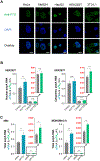
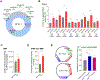
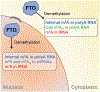
FTO, the first RNA demethylase discovered, mediates the demethylation of internal N6-methyladenosine (m6A) and N6, 2-O-dimethyladenosine (m6Am) at the +1 position from the 5' cap in mRNA. Here we demonstrate that the cellular distribution of FTO is distinct among different cell lines, affecting the access of FTO to different RNA substrates. We find that FTO binds multiple RNA species, including mRNA, snRNA, and tRNA, and can demethylate internal m6A and cap m6Am in mRNA, internal m6A in U6 RNA, internal and cap m6Am in snRNAs, and N1-methyladenosine (m1A) in tRNA. FTO-mediated demethylation has a greater effect on the transcript levels of mRNAs possessing internal m6A than the ones with cap m6Am in the tested cells. We also show that FTO can directly repress translation by catalyzing m1A tRNA demethylation. Collectively, FTO-mediated RNA demethylation occurs to m6A and m6Am in mRNA and snRNA as well as m1A in tRNA.
Keywords: FTO; cap m(6)A(m); cytoplasmic demethylation; m(6)A; nuclear m(6)A demethylation; snRNA demethylation; tRNA m(1)A demethylation; translation regulation.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
Chuan He is a scientific founder of Accent Therapeutics, Inc. and a member of its scientific advisory board.
Figures


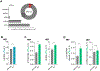

Similar articles
-
FTO controls reversible m6Am RNA methylation during snRNA biogenesis.Nat Chem Biol. 2019 Apr;15(4):340-347. doi: 10.1038/s41589-019-0231-8. Epub 2019 Feb 18. Nat Chem Biol. 2019. PMID: 30778204 Free PMC article.
-
Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates.Proc Natl Acad Sci U S A. 2019 Feb 19;116(8):2919-2924. doi: 10.1073/pnas.1820574116. Epub 2019 Feb 4. Proc Natl Acad Sci U S A. 2019. PMID: 30718435 Free PMC article.
-
FTO-mediated cytoplasmic m6Am demethylation adjusts stem-like properties in colorectal cancer cell.Nat Commun. 2021 Mar 19;12(1):1716. doi: 10.1038/s41467-021-21758-4. Nat Commun. 2021. PMID: 33741917 Free PMC article.
-
FTO: An Emerging Molecular Player in Neuropsychiatric Diseases.Neuroscience. 2019 Oct 15;418:15-24. doi: 10.1016/j.neuroscience.2019.08.021. Epub 2019 Aug 21. Neuroscience. 2019. PMID: 31442565 Review.
-
FTO, m6 Am , and the hypothesis of reversible epitranscriptomic mRNA modifications.FEBS Lett. 2018 Jun;592(12):2012-2022. doi: 10.1002/1873-3468.13092. Epub 2018 May 24. FEBS Lett. 2018. PMID: 29754392 Review.
Cited by
-
Sequencing methods and functional decoding of mRNA modifications.Fundam Res. 2023 Jun 1;3(5):738-748. doi: 10.1016/j.fmre.2023.05.010. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933299 Free PMC article. Review.
-
Gene Expression Profile and Prognostic Value of m6A RNA Methylation Regulators in Hepatocellular Carcinoma.J Hepatocell Carcinoma. 2021 Mar 12;8:85-101. doi: 10.2147/JHC.S296438. eCollection 2021. J Hepatocell Carcinoma. 2021. PMID: 33738268 Free PMC article.
-
Mechanisms of epitranscriptomic gene regulation.Biopolymers. 2021 Jan;112(1):e23403. doi: 10.1002/bip.23403. Epub 2020 Oct 1. Biopolymers. 2021. PMID: 33001446 Free PMC article. Review.
-
The Important Role of N6-methyladenosine RNA Modification in Non-Small Cell Lung Cancer.Genes (Basel). 2021 Mar 19;12(3):440. doi: 10.3390/genes12030440. Genes (Basel). 2021. PMID: 33808751 Free PMC article. Review.
-
RNA modifications in cardiovascular health and disease.Nat Rev Cardiol. 2023 May;20(5):325-346. doi: 10.1038/s41569-022-00804-8. Epub 2022 Dec 5. Nat Rev Cardiol. 2023. PMID: 36471164 Review.
References
-
- Adams JM, and Cory S (1975). Modified nucleosides and bizarre 5[prime]-termini in mouse myeloma mRNA. Nature 255, 28–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

