Quantitative Analysis of Alternative Pre-mRNA Splicing in Mouse Brain Sections Using RNA In Situ Hybridization Assay
- PMID: 30199013
- PMCID: PMC6231910
- DOI: 10.3791/57889
Quantitative Analysis of Alternative Pre-mRNA Splicing in Mouse Brain Sections Using RNA In Situ Hybridization Assay
Abstract
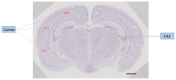
Alternative splicing (AS) occurs in more than 90% of human genes. The expression pattern of an alternatively spliced exon is often regulated in a cell type-specific fashion. AS expression patterns are typically analyzed by RT-PCR and RNA-seq using RNA samples isolated from a population of cells. In situ examination of AS expression patterns for a particular biological structure can be carried out by RNA in situ hybridization (ISH) using exon-specific probes. However, this particular use of ISH has been limited because alternative exons are generally too short to design exon-specific probes. In this report, the use of BaseScope, a recently developed technology that employs short antisense oligonucleotides in RNA ISH, is described to analyze AS expression patterns in mouse brain sections. Exon 23a of neurofibromatosis type 1 (Nf1) is used as an example to illustrate that short exon-exon junction probes exhibit robust hybridization signals with high specificity in RNA ISH analysis on mouse brain sections. More importantly, signals detected with exon inclusion- and skipping-specific probes can be used to reliably calculate the percent spliced in values of Nf1 exon 23a expression in different anatomical areas of a mouse brain. The experimental protocol and calculation method for AS analysis are presented. The results indicate that BaseScope provides a powerful new tool to assess AS expression patterns in situ.
Similar articles
-
A Novel Ultrasensitive In Situ Hybridization Approach to Detect Short Sequences and Splice Variants with Cellular Resolution.Mol Neurobiol. 2018 Jul;55(7):6169-6181. doi: 10.1007/s12035-017-0834-6. Epub 2017 Dec 20. Mol Neurobiol. 2018. PMID: 29264769 Free PMC article.
-
Alternative splicing of the neurofibromatosis type 1 pre-mRNA is regulated by the muscleblind-like proteins and the CUG-BP and ELAV-like factors.BMC Mol Biol. 2012 Dec 10;13:35. doi: 10.1186/1471-2199-13-35. BMC Mol Biol. 2012. PMID: 23227900 Free PMC article.
-
Alternative splicing of the neurofibromatosis type I pre-mRNA.Biosci Rep. 2012 Apr 1;32(2):131-8. doi: 10.1042/BSR20110060. Biosci Rep. 2012. PMID: 22115364 Free PMC article. Review.
-
Functional analysis of splicing mutations in exon 7 of NF1 gene.BMC Med Genet. 2007 Feb 12;8:4. doi: 10.1186/1471-2350-8-4. BMC Med Genet. 2007. PMID: 17295913 Free PMC article.
-
Invention and Early History of Exon Skipping and Splice Modulation.Methods Mol Biol. 2018;1828:3-30. doi: 10.1007/978-1-4939-8651-4_1. Methods Mol Biol. 2018. PMID: 30171532 Review.
Cited by
-
Circ-Hdgfrp3 shuttles along neurites and is trapped in aggregates formed by ALS-associated mutant FUS.iScience. 2021 Nov 25;24(12):103504. doi: 10.1016/j.isci.2021.103504. eCollection 2021 Dec 17. iScience. 2021. PMID: 34934923 Free PMC article.
-
Using antisense oligonucleotides for the physiological modulation of the alternative splicing of NF1 exon 23a during PC12 neuronal differentiation.Sci Rep. 2021 Feb 11;11(1):3661. doi: 10.1038/s41598-021-83152-w. Sci Rep. 2021. PMID: 33574490 Free PMC article.
-
Detection and Quantification of Multiple RNA Sequences Using Emerging Ultrasensitive Fluorescent In Situ Hybridization Techniques.Curr Protoc Neurosci. 2019 Apr;87(1):e63. doi: 10.1002/cpns.63. Epub 2019 Feb 21. Curr Protoc Neurosci. 2019. PMID: 30791216 Free PMC article.
References
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40:1413–1415. - PubMed
-
- Shirai Y, Watanabe M, Sakagami H, Suzuki T. Novel splice variants in the 5'UTR of Gtf2i expressed in the rat brain: alternative 5'UTRs and differential expression in the neuronal dendrites. Journal of Neurochemistry. 2015;134:578–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous