Barcoding of Plant Viruses with Circular Single-Stranded DNA Based on Rolling Circle Amplification
- PMID: 30200312
- PMCID: PMC6164888
- DOI: 10.3390/v10090469
Barcoding of Plant Viruses with Circular Single-Stranded DNA Based on Rolling Circle Amplification
Abstract
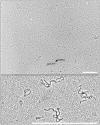
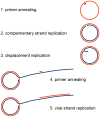
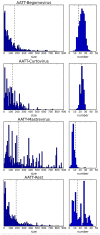
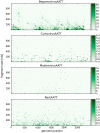
The experience with a diagnostic technology based on rolling circle amplification (RCA), restriction fragment length polymorphism (RFLP) analyses, and direct or deep sequencing (Circomics) over the past 15 years is surveyed for the plant infecting geminiviruses, nanoviruses and associated satellite DNAs, which have had increasing impact on agricultural and horticultural losses due to global transportation and recombination-aided diversification. Current state methods for quarantine measures are described to identify individual DNA components with great accuracy and to recognize the crucial role of the molecular viral population structure as an important factor for sustainable plant protection.
Keywords: Circomics; RCA; RFLP; geminivirus; nanovirus; satellite.
Conflict of interest statement
The author declares no conflict of interest.
Figures

Similar articles
-
Circomics of Cuban geminiviruses reveals the first alpha-satellite DNA in the Caribbean.Virus Genes. 2014 Oct;49(2):312-24. doi: 10.1007/s11262-014-1090-8. Epub 2014 Jun 19. Virus Genes. 2014. PMID: 24943118
-
Analysis of DNAs associated with coconut foliar decay disease implicates a unique single-stranded DNA virus representing a new taxon.Sci Rep. 2018 Apr 9;8(1):5698. doi: 10.1038/s41598-018-23739-y. Sci Rep. 2018. PMID: 29632309 Free PMC article.
-
Circular DNA genomics (circomics) exemplified for geminiviruses in bean crops and weeds of northeastern Brazil.Virology. 2012 Jun 5;427(2):151-7. doi: 10.1016/j.virol.2012.02.007. Epub 2012 Mar 6. Virology. 2012. PMID: 22397740
-
A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics.Arch Virol. 2012 Oct;157(10):1851-71. doi: 10.1007/s00705-012-1391-y. Epub 2012 Jul 4. Arch Virol. 2012. PMID: 22760663 Review.
-
Subviral agents associated with plant single-stranded DNA viruses.Virology. 2006 Jan 5;344(1):198-210. doi: 10.1016/j.virol.2005.09.042. Virology. 2006. PMID: 16364750 Review.
Cited by
-
A New Method to Obtain the Complete Genome Sequence of Multiple-Component Circular ssDNA Viruses by Transcriptome Analysis.Front Bioeng Biotechnol. 2020 Jul 21;8:832. doi: 10.3389/fbioe.2020.00832. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32850712 Free PMC article.
-
From Metagenomics to Ecogenomics: NGS-Based Approaches for Discovery of New Circular DNA Single-Stranded Viral Species.Methods Mol Biol. 2024;2732:103-117. doi: 10.1007/978-1-0716-3515-5_7. Methods Mol Biol. 2024. PMID: 38060120
-
Comparison of the gut virus communities between patients with Crohn's disease and healthy individuals.Front Microbiol. 2023 Jun 15;14:1190172. doi: 10.3389/fmicb.2023.1190172. eCollection 2023. Front Microbiol. 2023. PMID: 37396350 Free PMC article.
-
Phosphorylations of the Abutilon Mosaic Virus Movement Protein Affect Its Self-Interaction, Symptom Development, Viral DNA Accumulation, and Host Range.Front Plant Sci. 2020 Jul 31;11:1155. doi: 10.3389/fpls.2020.01155. eCollection 2020. Front Plant Sci. 2020. PMID: 32849713 Free PMC article.
-
Nanovirus Disease Complexes: An Emerging Threat in the Modern Era.Front Plant Sci. 2020 Nov 19;11:558403. doi: 10.3389/fpls.2020.558403. eCollection 2020. Front Plant Sci. 2020. PMID: 33329624 Free PMC article. Review.
References
-
- Moffat A. Geminiviruses emerge as serious crop threat. Science. 1999;286:1835. doi: 10.1126/science.286.5446.1835. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

