Lineage tracing of axial progenitors using Nkx1-2CreERT2 mice defines their trunk and tail contributions
- PMID: 30201686
- PMCID: PMC6198475
- DOI: 10.1242/dev.164319
Lineage tracing of axial progenitors using Nkx1-2CreERT2 mice defines their trunk and tail contributions
Abstract
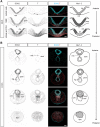
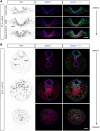
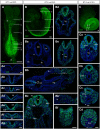
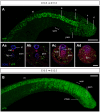
The vertebrate body forms by continuous generation of new tissue from progenitors at the posterior end of the embryo. The study of these axial progenitors has proved to be challenging in vivo largely because of the lack of unique molecular markers to identify them. Here, we elucidate the expression pattern of the transcription factor Nkx1-2 in the mouse embryo and show that it identifies axial progenitors throughout body axis elongation, including neuromesodermal progenitors and early neural and mesodermal progenitors. We create a tamoxifen-inducible Nkx1-2CreERT2 transgenic mouse and exploit the conditional nature of this line to uncover the lineage contributions of Nkx1-2-expressing cells at specific stages. We show that early Nkx1-2-expressing epiblast cells contribute to all three germ layers, mostly neuroectoderm and mesoderm, excluding notochord. Our data are consistent with the presence of some self-renewing axial progenitors that continue to generate neural and mesoderm tissues from the tail bud. This study identifies Nkx1-2-expressing cells as the source of most trunk and tail tissues in the mouse and provides a useful tool to genetically label and manipulate axial progenitors in vivo.
Keywords: Axial progenitors; Body axis elongation; Genetic lineage tracing; Mouse embryo; Neuromesodermal progenitors; Nkx1-2.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
-
- Beddington R. S. (1994). Induction of a second neural axis by the mouse node. Development 120, 613-620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
