Structure-specific DNA replication-fork recognition directs helicase and replication restart activities of the PriA helicase
- PMID: 30201718
- PMCID: PMC6166810
- DOI: 10.1073/pnas.1809842115
Structure-specific DNA replication-fork recognition directs helicase and replication restart activities of the PriA helicase
Abstract
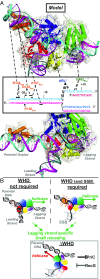
DNA replication restart, the essential process that reinitiates prematurely terminated genome replication reactions, relies on exquisitely specific recognition of abandoned DNA replication-fork structures. The PriA DNA helicase mediates this process in bacteria through mechanisms that remain poorly defined. We report the crystal structure of a PriA/replication-fork complex, which resolves leading-strand duplex DNA bound to the protein. Interaction with PriA unpairs one end of the DNA and sequesters the 3'-most nucleotide from the nascent leading strand into a conserved protein pocket. Cross-linking studies reveal a surface on the winged-helix domain of PriA that binds to parental duplex DNA. Deleting the winged-helix domain alters PriA's structure-specific DNA unwinding properties and impairs its activity in vivo. Our observations lead to a model in which coordinated parental-, leading-, and lagging-strand DNA binding provide PriA with the structural specificity needed to act on abandoned DNA replication forks.
Keywords: DNA repair; DNA replication restart; X-ray crystallography; cross-link mapping; protein–DNA complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cox MM, et al. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. - PubMed
-
- Jezewska MJ, Bujalowski W. Interactions of Escherichia coli replicative helicase PriA protein with single-stranded DNA. Biochemistry. 2000;39:10454–10467. - PubMed
-
- Szymanski MR, Jezewska MJ, Bujalowski W. The Escherichia coli PriA helicase-double-stranded DNA complex: Location of the strong DNA-binding subsite on the helicase domain of the protein and the affinity control by the two nucleotide-binding sites of the enzyme. J Mol Biol. 2010;402:344–362. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

