Function and crystal structure of the dimeric P-loop ATPase CFD1 coordinating an exposed [4Fe-4S] cluster for transfer to apoproteins
- PMID: 30201724
- PMCID: PMC6166825
- DOI: 10.1073/pnas.1807762115
Function and crystal structure of the dimeric P-loop ATPase CFD1 coordinating an exposed [4Fe-4S] cluster for transfer to apoproteins
Abstract
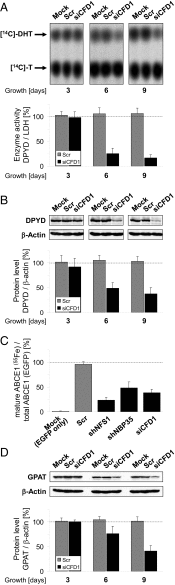
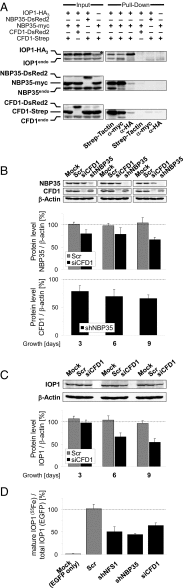
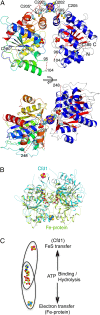
Maturation of iron-sulfur (Fe-S) proteins in eukaryotes requires complex machineries in mitochondria and cytosol. Initially, Fe-S clusters are assembled on dedicated scaffold proteins and then are trafficked to target apoproteins. Within the cytosolic Fe-S protein assembly (CIA) machinery, the conserved P-loop nucleoside triphosphatase Nbp35 performs a scaffold function. In yeast, Nbp35 cooperates with the related Cfd1, which is evolutionary less conserved and is absent in plants. Here, we investigated the potential scaffold function of human CFD1 (NUBP2) in CFD1-depleted HeLa cells by measuring Fe-S enzyme activities or 55Fe incorporation into Fe-S target proteins. We show that CFD1, in complex with NBP35 (NUBP1), performs a crucial role in the maturation of all tested cytosolic and nuclear Fe-S proteins, including essential ones involved in protein translation and DNA maintenance. CFD1 also matures iron regulatory protein 1 and thus is critical for cellular iron homeostasis. To better understand the scaffold function of CFD1-NBP35, we resolved the crystal structure of Chaetomium thermophilum holo-Cfd1 (ctCfd1) at 2.6-Å resolution as a model Cfd1 protein. Importantly, two ctCfd1 monomers coordinate a bridging [4Fe-4S] cluster via two conserved cysteine residues. The surface-exposed topology of the cluster is ideally suited for both de novo assembly and facile transfer to Fe-S apoproteins mediated by other CIA factors. ctCfd1 specifically interacted with ATP, which presumably associates with a pocket near the Cfd1 dimer interface formed by the conserved Walker motif. In contrast, ctNbp35 preferentially bound GTP, implying differential regulation of the two fungal scaffold components during Fe-S cluster assembly and/or release.
Keywords: CIA machinery; NBP35; NUBP1-NUBP2; iron homeostasis; iron-sulfur protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
A bridging [4Fe-4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron-sulfur protein maturation.J Biol Chem. 2012 Apr 6;287(15):12365-78. doi: 10.1074/jbc.M111.328914. Epub 2012 Feb 23. J Biol Chem. 2012. PMID: 22362766 Free PMC article.
-
The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol.Nat Chem Biol. 2007 May;3(5):278-86. doi: 10.1038/nchembio872. Epub 2007 Apr 1. Nat Chem Biol. 2007. PMID: 17401378
-
The conserved protein Dre2 uses essential [2Fe-2S] and [4Fe-4S] clusters for its function in cytosolic iron-sulfur protein assembly.Biochem J. 2016 Jul 15;473(14):2073-85. doi: 10.1042/BCJ20160416. Epub 2016 May 10. Biochem J. 2016. PMID: 27166425
-
Mechanisms of iron-sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes.Biochim Biophys Acta. 2006 Jul;1763(7):652-67. doi: 10.1016/j.bbamcr.2006.05.011. Epub 2006 May 23. Biochim Biophys Acta. 2006. PMID: 16843540 Review.
-
Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis.Annu Rev Biochem. 2020 Jun 20;89:471-499. doi: 10.1146/annurev-biochem-013118-111540. Epub 2020 Jan 14. Annu Rev Biochem. 2020. PMID: 31935115 Review.
Cited by
-
Nubp2 is required for cranial neural crest survival in the mouse.Dev Biol. 2020 Feb 15;458(2):189-199. doi: 10.1016/j.ydbio.2019.10.039. Epub 2019 Nov 14. Dev Biol. 2020. PMID: 31733190 Free PMC article.
-
Abnormal upregulation of NUBP2 contributes to cancer progression in colorectal cancer.Mol Cell Biochem. 2025 Jan;480(1):399-410. doi: 10.1007/s11010-024-04956-8. Epub 2024 Mar 16. Mol Cell Biochem. 2025. PMID: 38492158 Free PMC article.
-
Sulfur Administration in Fe-S Cluster Homeostasis.Antioxidants (Basel). 2021 Oct 29;10(11):1738. doi: 10.3390/antiox10111738. Antioxidants (Basel). 2021. PMID: 34829609 Free PMC article. Review.
-
Crucial role and conservation of the three [2Fe-2S] clusters in the human mitochondrial ribosome.J Biol Chem. 2025 Feb;301(2):108087. doi: 10.1016/j.jbc.2024.108087. Epub 2024 Dec 13. J Biol Chem. 2025. PMID: 39675708 Free PMC article.
-
Ancestral [Fe-S] biogenesis system SMS has a unique mechanism of cluster assembly and sulfur utilization.PLoS Biol. 2025 Jun 25;23(6):e3003223. doi: 10.1371/journal.pbio.3003223. eCollection 2025 Jun. PLoS Biol. 2025. PMID: 40560930 Free PMC article.
References
-
- Saha PP, Vishwanathan V, Bankapalli K, D’Silva P. Iron-sulfur protein assembly in human cells. Rev Physiol Biochem Pharmacol. 2017;174:25–65. - PubMed
-
- Ciofi-Baffoni S, Nasta V, Banci L. Protein networks in the maturation of human iron-sulfur proteins. Metallomics. 2018;10:49–72. - PubMed
-
- Lill R, Srinivasan V, Mühlenhoff U. The role of mitochondria in cytosolic-nuclear iron–sulfur protein biogenesis and in cellular iron regulation. Curr Opin Microbiol. 2014;22:111–119. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

