Comparative Secretome Analyses of Human and Zoonotic Staphylococcus aureus Isolates CC8, CC22, and CC398
- PMID: 30201737
- PMCID: PMC6283302
- DOI: 10.1074/mcp.RA118.001036
Comparative Secretome Analyses of Human and Zoonotic Staphylococcus aureus Isolates CC8, CC22, and CC398
Abstract
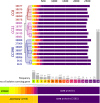
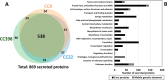
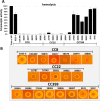
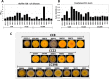
The spread of methicillin-resistant Staphylococcus aureus (MRSA) in the community, hospitals and in livestock is mediated by highly diverse virulence factors that include secreted toxins, superantigens, enzymes and surface-associated adhesins allowing host adaptation and colonization. Here, we combined proteogenomics, secretome and phenotype analyses to compare the secreted virulence factors in selected S. aureus isolates of the dominant human- and livestock-associated genetic lineages CC8, CC22, and CC398. The proteogenomic comparison revealed 2181 core genes and 1306 accessory genes in 18 S. aureus isolates reflecting the high genome diversity. Using secretome analysis, we identified 869 secreted proteins with 538 commons in eight isolates of CC8, CC22, and CC398. These include 64 predicted extracellular and 37 cell surface proteins that account for 82.4% of total secretome abundance. Among the top 10 most abundantly secreted virulence factors are the major autolysins (Atl, IsaA, Sle1, SAUPAN006375000), lipases and lipoteichoic acid hydrolases (Lip, Geh, LtaS), cytolytic toxins (Hla, Hlb, PSMβ1) and proteases (SspB). The CC398 isolates showed lower secretion of cell wall proteins, but higher secretion of α- and β-hemolysins (Hla, Hlb) which correlated with an increased Agr activity and strong hemolysis. CC398 strains were further characterized by lower biofilm formation and staphyloxanthin levels because of decreased SigB activity. Overall, comparative secretome analyses revealed CC8- or CC22-specific enterotoxin and Spl protease secretion as well as Agr- and SigB-controlled differences in exotoxin and surface protein secretion between human-specific and zoonotic lineages of S. aureus.
Keywords: MRSA; Microbiology; Pathogens; Proteogenomics; Secretome; Staphylococcus aureus; Virulence; clonal complexes; virulence factor secretion.
© 2018 Busche et al.
Figures






Similar articles
-
Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock.mBio. 2012 Feb 21;3(1):e00305-11. doi: 10.1128/mBio.00305-11. Print 2012. mBio. 2012. PMID: 22354957 Free PMC article.
-
LA-MRSA CC398 differ from classical community acquired-MRSA and hospital acquired-MRSA lineages: functional analysis of infection and colonization processes.Int J Med Microbiol. 2014 Oct;304(7):777-86. doi: 10.1016/j.ijmm.2014.06.006. Epub 2014 Jun 27. Int J Med Microbiol. 2014. PMID: 25034858
-
Livestock associated Staphylococcus aureus in cystic fibrosis patients in Spain: Detection of MRSA and MSSA CC398.Microb Pathog. 2024 Dec;197:107016. doi: 10.1016/j.micpath.2024.107016. Epub 2024 Oct 15. Microb Pathog. 2024. PMID: 39413853
-
Comparative review of the nasal carriage and genetic characteristics of Staphylococcus aureus in healthy livestock: Insight into zoonotic and anthroponotic clones.Infect Genet Evol. 2023 Apr;109:105408. doi: 10.1016/j.meegid.2023.105408. Epub 2023 Feb 10. Infect Genet Evol. 2023. PMID: 36773670 Review.
-
The pathogenicity and host adaptation of livestock-associated MRSA CC398.Vet Microbiol. 2017 Feb;200:39-45. doi: 10.1016/j.vetmic.2016.05.006. Epub 2016 May 14. Vet Microbiol. 2017. PMID: 27236228 Review.
Cited by
-
How to survive pig farming: Mechanism of SCCmec element deletion and metabolic stress adaptation in livestock-associated MRSA.Front Microbiol. 2022 Nov 23;13:969961. doi: 10.3389/fmicb.2022.969961. eCollection 2022. Front Microbiol. 2022. PMID: 36504815 Free PMC article.
-
Comparative Secretome Analysis of Magnaporthe oryzae Identified Proteins Involved in Virulence and Cell Wall Integrity.Genomics Proteomics Bioinformatics. 2022 Aug;20(4):728-746. doi: 10.1016/j.gpb.2021.02.007. Epub 2021 Jul 18. Genomics Proteomics Bioinformatics. 2022. PMID: 34284133 Free PMC article.
-
Micrococcal nuclease regulates biofilm formation and dispersal in methicillin-resistant Staphylococcus aureus USA300.bioRxiv [Preprint]. 2023 Nov 5:2023.11.05.565664. doi: 10.1101/2023.11.05.565664. bioRxiv. 2023. Update in: mSphere. 2024 May 29;9(5):e0012624. doi: 10.1128/msphere.00126-24. PMID: 37961602 Free PMC article. Updated. Preprint.
-
Exoproteomic profiling uncovers critical determinants for virulence of livestock-associated and human-originated Staphylococcus aureus ST398 strains.Virulence. 2020 Dec;11(1):947-963. doi: 10.1080/21505594.2020.1793525. Virulence. 2020. PMID: 32726182 Free PMC article.
-
A Kayvirus Distant Homolog of Staphylococcal Virulence Determinants and VISA Biomarker Is a Phage Lytic Enzyme.Viruses. 2020 Mar 7;12(3):292. doi: 10.3390/v12030292. Viruses. 2020. PMID: 32156046 Free PMC article.
References
-
- Wertheim H. F. L., Melles D. C., Vos M. C., van Leeuwen W., van Belkum A., Verbrugh H. A., and Nouwen J. L. (2005) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762 - PubMed
-
- Weidenmaier C., Goerke C., and Wolz C. (2012) Staphylococcus aureus determinants for nasal colonization. Trends Microbiol. 20, 243–250 - PubMed
-
- Lowy F. D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 - PubMed
-
- Boucher H. W., and Corey G. R. (2008) Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46, S344–S349 - PubMed
-
- Archer G. L. (1998) Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26, 1179–1181 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

