CovR and VicRKX Regulate Transcription of the Collagen Binding Protein Cnm of Streptococcus mutans
- PMID: 30201780
- PMCID: PMC6222205
- DOI: 10.1128/JB.00141-18
CovR and VicRKX Regulate Transcription of the Collagen Binding Protein Cnm of Streptococcus mutans
Abstract
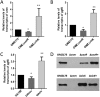
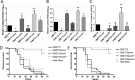
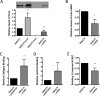
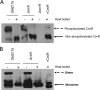
Cnm is a surface-associated protein present in a subset of Streptococcus mutans strains that mediates binding to extracellular matrices, intracellular invasion, and virulence. Here, we showed that cnm transcription is controlled by the global regulators CovR and VicRKX. In silico analysis identified multiple putative CovR- and VicR-binding motifs in the regulatory region of cnm as well as in the downstream gene pgfS, which is associated with the posttranslational modification of Cnm. Electrophoretic mobility shift assays revealed that CovR and VicR specifically and independently bind to the cnm and pgfS promoter regions. Quantitative real-time PCR and Western blot analyses of ΔcovR and ΔvicK strains as well as of a strain overexpressing vicRKX revealed that CovR functions as a positive regulator of cnm, whereas VicRKX acts as a negative regulator. In agreement with the role of VicRKX as a repressor, the ΔvicK strain showed enhanced binding to collagen and laminin and higher intracellular invasion rates. Overexpression of vicRKX was associated with decreased rates of intracellular invasion but did not affect collagen or lamin binding activities, suggesting that this system controls additional genes involved in binding to these extracellular matrix proteins. As expected, based on the role of CovR in cnm regulation, the ΔcovR strain showed decreased intracellular invasion rates, but, unexpectedly collagen and laminin binding activities were increased in this mutant strain. Collectively, the results presented here expand the repertoire of virulence-related genes regulated by CovR and VicRKX to include the core gene pgfS and the noncore gene cnmIMPORTANCEStreptococcus mutans is a major pathogen associated with dental caries and also implicated in systemic infections, in particular, infective endocarditis. The Cnm adhesin of S. mutans is an important virulence factor associated with systemic infections and caries severity. Despite its role in virulence, the regulatory mechanisms governing cnm expression are poorly understood. Here, we describe the identification of two independent regulatory systems controlling the transcription of cnm and the downstream pgfS-pgfM1-pgfE-pgfM2 operon. A better understanding of the mechanisms controlling expression of virulence factors like Cnm can facilitate the development of new strategies to treat bacterial infections.
Keywords: Streptococcus mutans; adhesins; host cell invasion; virulence regulation.
Copyright © 2018 American Society for Microbiology.
Figures

Similar articles
-
Characterization of the pgf operon involved in the posttranslational modification of Streptococcus mutans surface proteins.Sci Rep. 2018 Mar 16;8(1):4705. doi: 10.1038/s41598-018-23170-3. Sci Rep. 2018. PMID: 29549320 Free PMC article.
-
Cnm is a major virulence factor of invasive Streptococcus mutans and part of a conserved three-gene locus.Mol Oral Microbiol. 2014 Feb;29(1):11-23. doi: 10.1111/mom.12041. Epub 2013 Oct 30. Mol Oral Microbiol. 2014. PMID: 24103776 Free PMC article.
-
Modification of Streptococcus mutans Cnm by PgfS contributes to adhesion, endothelial cell invasion, and virulence.J Bacteriol. 2014 Aug;196(15):2789-97. doi: 10.1128/JB.01783-14. Epub 2014 May 16. J Bacteriol. 2014. PMID: 24837294 Free PMC article.
-
Collagen-binding proteins of Streptococcus mutans and related streptococci.Mol Oral Microbiol. 2017 Apr;32(2):89-106. doi: 10.1111/omi.12158. Epub 2016 Apr 25. Mol Oral Microbiol. 2017. PMID: 26991416 Free PMC article. Review.
-
The VicRK Two-Component System Regulates Streptococcus mutans Virulence.Curr Issues Mol Biol. 2019;32:167-200. doi: 10.21775/cimb.032.167. Epub 2019 Jun 5. Curr Issues Mol Biol. 2019. PMID: 31166172 Review.
Cited by
-
Post-translational modification by the Pgf glycosylation machinery modulates Streptococcus mutans OMZ175 physiology and virulence.Mol Microbiol. 2024 Aug;122(2):133-151. doi: 10.1111/mmi.15190. Epub 2023 Nov 16. Mol Microbiol. 2024. PMID: 37972006 Free PMC article.
-
Novel virulence factor Cba induces antibody-dependent enhancement (ADE) of Streptococcus suis Serotype 9 infection in a mouse model.Front Cell Infect Microbiol. 2023 Feb 21;13:1027419. doi: 10.3389/fcimb.2023.1027419. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36896190 Free PMC article.
-
A Case Series and Literature Review of Subacute Infective Endocarditis: A Clinical Challenge.Cureus. 2023 Mar 10;15(3):e35997. doi: 10.7759/cureus.35997. eCollection 2023 Mar. Cureus. 2023. PMID: 37041916 Free PMC article.
-
Potential involvement of Streptococcus mutans possessing collagen binding protein Cnm in infective endocarditis.Sci Rep. 2020 Nov 5;10(1):19118. doi: 10.1038/s41598-020-75933-6. Sci Rep. 2020. PMID: 33154489 Free PMC article.
-
Relationship between Streptococcus mutans expressing Cnm in the oral cavity and non-alcoholic steatohepatitis: a pilot study.BMJ Open Gastroenterol. 2019 Oct 1;6(1):e000329. doi: 10.1136/bmjgast-2019-000329. eCollection 2019. BMJ Open Gastroenterol. 2019. PMID: 31645988 Free PMC article.
References
-
- Nakano K, Nomura R, Taniguchi N, Lapirattanakul J, Kojima A, Naka S, Senawongse P, Srisatjaluk R, Gronroos L, Alaluusua S, Matsumoto M, Ooshima T. 2010. Molecular characterization of Streptococcus mutans strains containing the cnm gene encoding a collagen-binding adhesin. Arch Oral Biol 55:34–39. doi:10.1016/j.archoralbio.2009.11.008. - DOI - PubMed
-
- Abranches J, Miller JH, Martinez AR, Simpson-Haidaris PJ, Burne RA, Lemos JA. 2011. The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect Immun 79:2277–2284. doi:10.1128/IAI.00767-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

