Towards systematic, data-driven validation of a collaborative, multi-scale model of Caenorhabditis elegans
- PMID: 30201844
- PMCID: PMC6158230
- DOI: 10.1098/rstb.2017.0381
Towards systematic, data-driven validation of a collaborative, multi-scale model of Caenorhabditis elegans
Abstract
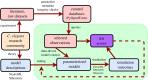
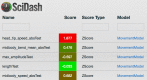
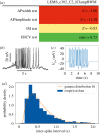
The OpenWorm Project is an international open-source collaboration to create a multi-scale model of the organism Caenorhabditis elegans At each scale, including subcellular, cellular, network and behaviour, this project employs one or more computational models that aim to recapitulate the corresponding biological system at that scale. This requires that the simulated behaviour of each model be compared with experimental data both as the model is continuously refined and as new experimental data become available. Here we report the use of SciUnit, a software framework for model validation, to attempt to achieve these goals. During project development, each model is continuously subjected to data-driven 'unit tests' that quantitatively summarize model-data agreement, identifying modelling progress and highlighting particular aspects of each model that fail to adequately reproduce known features of the biological organism and its components. This workflow is publicly visible via both GitHub and a web application and accepts community contributions to ensure that modelling goals are transparent and well-informed.This article is part of a discussion meeting issue 'Connectome to behaviour: modelling C. elegans at cellular resolution'.
Keywords: Python; informatics; modelling; unit-testing.
© 2018 The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Beck K. 2003. Test-driven development: by example. Boston, MA: Addison-Wesley Professional.
-
- Omar C, Aldrich J, Gerkin RC. 2014. Collaborative Infrastructure for Test-driven Scientific Model Validation. In Companion Proc. of the 36th International Conf. on Software Engineering, ICSE Companion 2014, pp. 524–527, New York, NY, USA: ACM; http://doi.acm.org/10.1145/2591062.2591129.
-
- Hines ML, Carnevale NT. 1997. The NEURON simulation environment. Neural Comput. 9, 1179–1209. - PubMed
-
- Yu Palyanov A, Khayrulin SS. 2015. Sibernetic: a software complex based on the PCI SPH algorithm aimed at simulation problems in biomechanics. Russ. J. Genet Appl. Res. 5, 635–641. ( 10.1134/S2079059715060052) - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
