Biogenic Polyphosphate Nanoparticles from a Marine Cyanobacterium Synechococcus sp. PCC 7002: Production, Characterization, and Anti-Inflammatory Properties In Vitro
- PMID: 30201855
- PMCID: PMC6163655
- DOI: 10.3390/md16090322
Biogenic Polyphosphate Nanoparticles from a Marine Cyanobacterium Synechococcus sp. PCC 7002: Production, Characterization, and Anti-Inflammatory Properties In Vitro
Abstract
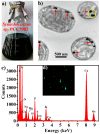
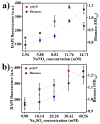
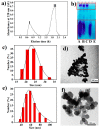
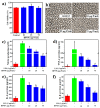
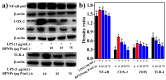
Probiotic-derived polyphosphates have attracted interest as potential therapeutic agents to improve intestinal health. The current study discovered the intracellular accumulation of polyphosphates in a marine cyanobacterium Synechococcus sp. PCC 7002 as nano-sized granules. The maximum accumulation of polyphosphates in Synechococcus sp. PCC 7002 was found at the late logarithmic growth phase when the medium contained 0.74 mM of KH₂PO₄, 11.76 mM of NaNO₃, and 30.42 mM of Na₂SO₄. Biogenic polyphosphate nanoparticles (BPNPs) were obtained intact from the algae cells by hot water extraction, and were purified to remove the organic impurities by Sephadex G-100 gel filtration. By using 100 kDa ultrafiltration, BPNPs were fractionated into the larger and smaller populations with diameters ranging between 30⁻70 nm and 10⁻30 nm, respectively. 4',6-diamidino-2-phenylindole fluorescence and orthophosphate production revealed that a minor portion of BPNPs (about 14⁻18%) were degraded during simulated gastrointestinal digestion. In vitro studies using lipopolysaccharide-activated RAW264.7 cells showed that BPNPs inhibited cyclooxygenase-2, inducible nitric oxide (NO) synthase expression, and the production of proinflammatory mediators, including NO, tumor necrosis factor-α, interleukin-6, and interleukin-1β through suppressing the Toll-like receptor 4/NF-κB signaling pathway. Overall, there is promise in the use of the marine cyanobacterium Synechococcus sp. PCC 7002 to produce BPNPs, an anti-inflammatory postbiotic.
Keywords: Synechococcus sp. PCC 7002; Toll-like receptors; anti-inflammation; macrophages; polyphosphate nanoparticles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Kulaev I.S., Vagabov V.M., Kulakovskaya T.V. The Biochemistry of Inorganic Polyphosphates. Springer International Publishing; Cham, Switzerland: 2005. pp. 3–35.
-
- Bonting C.F.C., Kortstee G.J.J., Boekestein A., Zehnder A.J. The elemental composition dynamics of large polyphosphate granules in Acinetobacter strain 210A. Arch. Microbiol. 1993;159:428–434. doi: 10.1007/BF00288589. - DOI
-
- Racki L.R., Tocheva E.I., Dieterle M.G., Sullivan M.C., Jensen G.J., Newman D.K. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2017;114:E2440–E2449. doi: 10.1073/pnas.1615575114. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

