An immunoregulatory and tissue-residency program modulated by c-MAF in human TH17 cells
- PMID: 30201991
- PMCID: PMC6402560
- DOI: 10.1038/s41590-018-0200-5
An immunoregulatory and tissue-residency program modulated by c-MAF in human TH17 cells
Erratum in
-
Publisher Correction: An immunoregulatory and tissue-residency program modulated by c-MAF in human TH17 cells.Nat Immunol. 2019 Jan;20(1):109. doi: 10.1038/s41590-018-0264-2. Nat Immunol. 2019. PMID: 30448856
Abstract
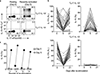
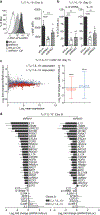
Different types of effector and memory T lymphocytes are induced and maintained in protective or pathological immune responses. Here we characterized two human CD4+ TH17 helper cell subsets that, in the recently activated state, could be distinguished on the basis of their expression of the anti-inflammatory cytokine IL-10. IL-10+ TH17 cells upregulated a variety of genes encoding immunoregulatory molecules, as well as genes whose expression is characteristic of tissue-resident T cells. In contrast, IL-10- TH17 cells maintained a pro-inflammatory gene-expression profile and upregulated the expression of homing receptors that guide recirculation from tissues to blood. Expression of the transcription factor c-MAF was selectively upregulated in IL-10+ TH17 cells, and it was bound to a large set of enhancer-like regions and modulated the immunoregulatory and tissue-residency program. Our results identify c-MAF as a relevant factor that drives two highly divergent post-activation fates of human TH17 cells and provide a framework with which to investigate the role of these cells in physiology and immunopathology.
Conflict of interest statement
Competing Financial Interests Statement
The authors declare no competing interests.
Figures
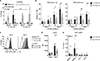




References
-
- Sallusto F, Lenig D, Forster R, Lipp M & Lanzavecchia A Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999). - PubMed
-
- Mueller SN & Mackay LK Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol 16, 79–89 (2016). - PubMed
-
- Ouyang W, Rutz S, Crellin NK, Valdez PA & Hymowitz SG Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol 29, 71–109 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

