Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota
- PMID: 30202015
- PMCID: PMC6179362
- DOI: 10.1038/s41564-018-0229-0
Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota
Abstract
As a conserved pathway that lies at the intersection between host defence and cellular homeostasis, autophagy serves as a rheostat for immune reactions. In particular, autophagy suppresses excess type I interferon (IFN-I) production in response to viral nucleic acids. It is unknown how this function of autophagy relates to the intestinal barrier where host-microbe interactions are pervasive and perpetual. Here, we demonstrate that mice deficient in autophagy proteins are protected from the intestinal bacterial pathogen Citrobacter rodentium in a manner dependent on IFN-I signalling and nucleic acid sensing pathways. Enhanced IFN-stimulated gene expression in intestinal tissue of autophagy-deficient mice in the absence of infection was mediated by the gut microbiota. Additionally, monocytes infiltrating into the autophagy-deficient intestinal microenvironment displayed an enhanced inflammatory profile and were necessary for protection against C. rodentium. Finally, we demonstrate that the microbiota-dependent IFN-I production that occurs in the autophagy-deficient host also protects against chemical injury of the intestine. Thus, autophagy proteins prevent a spontaneous IFN-I response to microbiota that is beneficial in the presence of infectious and non-infectious intestinal hazards. These results identify a role for autophagy proteins in controlling the magnitude of IFN-I signalling at the intestinal barrier.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
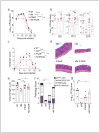
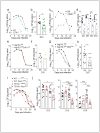
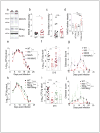

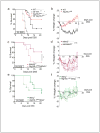

Comment in
-
Taming the beasts within.Nat Microbiol. 2018 Oct;3(10):1080-1081. doi: 10.1038/s41564-018-0264-x. Nat Microbiol. 2018. PMID: 30254330 No abstract available.
Similar articles
-
New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD.Autophagy. 2020 Jan;16(1):38-51. doi: 10.1080/15548627.2019.1635384. Epub 2019 Jul 9. Autophagy. 2020. PMID: 31286804 Free PMC article. Review.
-
Inhibition of Atg7 in intestinal epithelial cells drives resistance against Citrobacter rodentium.Cell Death Dis. 2025 Feb 19;16(1):112. doi: 10.1038/s41419-025-07422-5. Cell Death Dis. 2025. PMID: 39971913 Free PMC article.
-
IFN-I and IL-22 mediate protective effects of intestinal viral infection.Nat Microbiol. 2019 Oct;4(10):1737-1749. doi: 10.1038/s41564-019-0470-1. Epub 2019 Jun 10. Nat Microbiol. 2019. PMID: 31182797 Free PMC article.
-
Regulation of interferon signaling in response to gut microbes by autophagy.Gut Microbes. 2020;11(1):126-134. doi: 10.1080/19490976.2019.1614395. Epub 2019 May 23. Gut Microbes. 2020. PMID: 31119977 Free PMC article.
-
Overview of the Effect of Citrobacter rodentium Infection on Host Metabolism and the Microbiota.Methods Mol Biol. 2021;2291:399-418. doi: 10.1007/978-1-0716-1339-9_20. Methods Mol Biol. 2021. PMID: 33704766 Review.
Cited by
-
IL-17RA-signaling in Lgr5+ intestinal stem cells induces expression of transcription factor ATOH1 to promote secretory cell lineage commitment.Immunity. 2022 Feb 8;55(2):237-253.e8. doi: 10.1016/j.immuni.2021.12.016. Epub 2022 Jan 25. Immunity. 2022. PMID: 35081371 Free PMC article.
-
New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD.Autophagy. 2020 Jan;16(1):38-51. doi: 10.1080/15548627.2019.1635384. Epub 2019 Jul 9. Autophagy. 2020. PMID: 31286804 Free PMC article. Review.
-
Shared and Distinct Functions of Type I and Type III Interferons.Immunity. 2019 Apr 16;50(4):907-923. doi: 10.1016/j.immuni.2019.03.025. Immunity. 2019. PMID: 30995506 Free PMC article. Review.
-
Dying by fire: noncanonical functions of autophagy proteins in neuroinflammation and neurodegeneration.Neural Regen Res. 2022 Feb;17(2):246-250. doi: 10.4103/1673-5374.317958. Neural Regen Res. 2022. PMID: 34269183 Free PMC article. Review.
-
Inhibition of Atg7 in intestinal epithelial cells drives resistance against Citrobacter rodentium.Cell Death Dis. 2025 Feb 19;16(1):112. doi: 10.1038/s41419-025-07422-5. Cell Death Dis. 2025. PMID: 39971913 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

