Characterisation of pathogen-specific regions and novel effector candidates in Fusarium oxysporum f. sp. cepae
- PMID: 30202022
- PMCID: PMC6131394
- DOI: 10.1038/s41598-018-30335-7
Characterisation of pathogen-specific regions and novel effector candidates in Fusarium oxysporum f. sp. cepae
Abstract
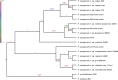
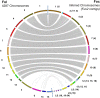
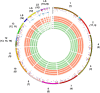
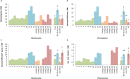
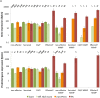
A reference-quality assembly of Fusarium oxysporum f. sp. cepae (Foc), the causative agent of onion basal rot has been generated along with genomes of additional pathogenic and non-pathogenic isolates of onion. Phylogenetic analysis confirmed a single origin of the Foc pathogenic lineage. Genome alignments with other F. oxysporum ff. spp. and non pathogens revealed high levels of syntenic conservation of core chromosomes but little synteny between lineage specific (LS) chromosomes. Four LS contigs in Foc totaling 3.9 Mb were designated as pathogen-specific (PS). A two-fold increase in segmental duplication events was observed between LS regions of the genome compared to within core regions or from LS regions to the core. RNA-seq expression studies identified candidate effectors expressed in planta, consisting of both known effector homologs and novel candidates. FTF1 and a subset of other transcription factors implicated in regulation of effector expression were found to be expressed in planta.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Leslie, J. F. & Summerell, B. A. The Fusarium Laboratory Manual. (Oxford: Blackwell Publishing, 2006).
Publication types
MeSH terms
Substances
Grants and funding
- BB/K020730/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020870/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020730/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020870a/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020730/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020730/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020730/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020870a/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020870a/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020730/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/K020870a/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

