PoreDesigner for tuning solute selectivity in a robust and highly permeable outer membrane pore
- PMID: 30202038
- PMCID: PMC6131167
- DOI: 10.1038/s41467-018-06097-1
PoreDesigner for tuning solute selectivity in a robust and highly permeable outer membrane pore
Abstract
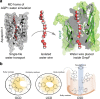
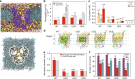
Monodispersed angstrom-size pores embedded in a suitable matrix are promising for highly selective membrane-based separations. They can provide substantial energy savings in water treatment and small molecule bioseparations. Such pores present as membrane proteins (chiefly aquaporin-based) are commonplace in biological membranes but difficult to implement in synthetic industrial membranes and have modest selectivity without tunable selectivity. Here we present PoreDesigner, a design workflow to redesign the robust beta-barrel Outer Membrane Protein F as a scaffold to access three specific pore designs that exclude solutes larger than sucrose (>360 Da), glucose (>180 Da), and salt (>58 Da) respectively. PoreDesigner also enables us to design any specified pore size (spanning 3-10 Å), engineer its pore profile, and chemistry. These redesigned pores may be ideal for conducting sub-nm aqueous separations with permeabilities exceeding those of classical biological water channels, aquaporins, by more than an order of magnitude at over 10 billion water molecules per channel per second.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Levin, R. J. The Living Barrier: A Primer on Transfer Across Biological Membranes (Butterworth-Heinemann, London, UK, 2014).
-
- Grzelakowski M, Cherenet MF, Shen YX, Kumar M. A framework for accurate evaluation of the promise of aquaporin based biomimetic membranes. J. Memb. Sci. 2015;479:223–231. doi: 10.1016/j.memsci.2015.01.023. - DOI
-
- Crittenden, J. C., Trussell, R. R., Hand, D. W., Howe, K. J. & Tchobanoglous, G. MWH’s Water Treatment: Principles and Design 3rd edn (John Wiley & Sons, Inc., 2012).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

