Crystal structure of the modification-dependent SRA-HNH endonuclease TagI
- PMID: 30202937
- PMCID: PMC6212794
- DOI: 10.1093/nar/gky781
Crystal structure of the modification-dependent SRA-HNH endonuclease TagI
Abstract
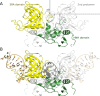
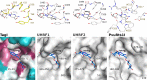
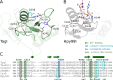
TagI belongs to the recently characterized SRA-HNH family of modification-dependent restriction endonucleases (REases) that also includes ScoA3IV (Sco5333) and TbiR51I (Tbis1). Here, we present a crystal structure of dimeric TagI, which exhibits a DNA binding site formed jointly by the nuclease domains, and separate binding sites for modified DNA bases in the two protomers. The nuclease domains have characteristic features of HNH/ββα-Me REases, and catalyze nicks or double strand breaks, with preference for /RY and RYN/RY sites, respectively. The SRA domains have the canonical fold. Their pockets for the flipped bases are spacious enough to accommodate 5-methylcytosine (5mC) or 5-hydroxymethylcytosine (5hmC), but not glucosyl-5-hydroxymethylcytosine (g5hmC). Such preference is in agreement with the biochemical determination of the TagI modification dependence and the results of phage restriction assays. The ability of TagI to digest plasmids methylated by Dcm (C5mCWGG), M.Fnu4HI (G5mCNGC) or M.HpyCH4IV (A5mCGT) suggests that the SRA domains of the enzyme are tolerant to different sequence contexts of the modified base.
Figures

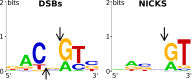

Similar articles
-
Recognition and cleavage of 5-methylcytosine DNA by bacterial SRA-HNH proteins.Nucleic Acids Res. 2015 Jan;43(2):1147-59. doi: 10.1093/nar/gku1376. Epub 2015 Jan 6. Nucleic Acids Res. 2015. PMID: 25564526 Free PMC article.
-
Crystal structures of the EVE-HNH endonuclease VcaM4I in the presence and absence of DNA.Nucleic Acids Res. 2021 Feb 22;49(3):1708-1723. doi: 10.1093/nar/gkaa1218. Nucleic Acids Res. 2021. PMID: 33450012 Free PMC article.
-
Modification-dependent restriction endonuclease, MspJI, flips 5-methylcytosine out of the DNA helix.Nucleic Acids Res. 2014 Oct 29;42(19):12092-101. doi: 10.1093/nar/gku871. Epub 2014 Sep 27. Nucleic Acids Res. 2014. PMID: 25262349 Free PMC article.
-
Epigenetic Modifications of Cytosine: Biophysical Properties, Regulation, and Function in Mammalian DNA.Bioessays. 2018 Mar;40(3). doi: 10.1002/bies.201700199. Epub 2018 Jan 25. Bioessays. 2018. PMID: 29369386 Review.
-
Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the "midnight zone" of homology.Curr Protein Pept Sci. 2003 Oct;4(5):327-37. doi: 10.2174/1389203033487072. Curr Protein Pept Sci. 2003. PMID: 14529527 Review.
Cited by
-
Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.Nucleic Acids Res. 2019 Dec 16;47(22):11943-11955. doi: 10.1093/nar/gkz1017. Nucleic Acids Res. 2019. PMID: 31724709 Free PMC article.
-
HK97 gp74 Possesses an α-Helical Insertion in the ββα Fold That Affects Its Metal Binding, cos Site Digestion, and In Vivo Activities.J Bacteriol. 2020 Mar 26;202(8):e00644-19. doi: 10.1128/JB.00644-19. Print 2020 Mar 26. J Bacteriol. 2020. PMID: 31988081 Free PMC article.
-
A protein architecture guided screen for modification dependent restriction endonucleases.Nucleic Acids Res. 2019 Oct 10;47(18):9761-9776. doi: 10.1093/nar/gkz755. Nucleic Acids Res. 2019. PMID: 31504772 Free PMC article.
-
A Unique m6A-Dependent Restriction Endonuclease from an Archaeal Virus.Microbiol Spectr. 2023 Mar 22;11(2):e0426222. doi: 10.1128/spectrum.04262-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36946751 Free PMC article.
-
Structural analysis of the BisI family of modification dependent restriction endonucleases.Nucleic Acids Res. 2024 Aug 27;52(15):9103-9118. doi: 10.1093/nar/gkae634. Nucleic Acids Res. 2024. PMID: 39041409 Free PMC article.
References
-
- Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M.. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008; 455:818–821. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

