Dot1 promotes H2B ubiquitination by a methyltransferase-independent mechanism
- PMID: 30203048
- PMCID: PMC6265471
- DOI: 10.1093/nar/gky801
Dot1 promotes H2B ubiquitination by a methyltransferase-independent mechanism
Abstract
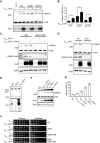
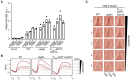
The histone methyltransferase Dot1 is conserved from yeast to human and methylates lysine 79 of histone H3 (H3K79) on the core of the nucleosome. H3K79 methylation by Dot1 affects gene expression and the response to DNA damage, and is enhanced by monoubiquitination of the C-terminus of histone H2B (H2Bub1). To gain more insight into the functions of Dot1, we generated genetic interaction maps of increased-dosage alleles of DOT1. We identified a functional relationship between increased Dot1 dosage and loss of the DUB module of the SAGA co-activator complex, which deubiquitinates H2Bub1 and thereby negatively regulates H3K79 methylation. Increased Dot1 dosage was found to promote H2Bub1 in a dose-dependent manner and this was exacerbated by the loss of SAGA-DUB activity, which also caused a negative genetic interaction. The stimulatory effect on H2B ubiquitination was mediated by the N-terminus of Dot1, independent of methyltransferase activity. Our findings show that Dot1 and H2Bub1 are subject to bi-directional crosstalk and that Dot1 possesses chromatin regulatory functions that are independent of its methyltransferase activity.
Figures


References
-
- Venkatesh S., Workman J.L.. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015; 16:178–189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

