Advances in mass spectrometry-based glycoproteomics
- PMID: 30203847
- PMCID: PMC6375712
- DOI: 10.1002/elps.201800272
Advances in mass spectrometry-based glycoproteomics
Abstract
Protein glycosylation, an important PTM, plays an essential role in a wide range of biological processes such as immune response, intercellular signaling, inflammation, and host-pathogen interaction. Aberrant glycosylation has been correlated with various diseases. However, studying protein glycosylation remains challenging because of low abundance, microheterogeneities of glycosylation sites, and poor ionization efficiency of glycopeptides. Therefore, the development of sensitive and accurate approaches to characterize protein glycosylation is crucial. The identification and characterization of protein glycosylation by MS is referred to as the field of glycoproteomics. Methods such as enrichment, metabolic labeling, and derivatization of glycopeptides in conjunction with different MS techniques and bioinformatics tools, have been developed to achieve an unequivocal quantitative and qualitative characterization of glycoproteins. This review summarizes the recent developments in the field of glycoproteomics over the past 6 years (2012 to 2018).
Keywords: Derivatization; Enrichment; Glycoproteins; Glycosylation; Metabolic labeling.
© 2018 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures

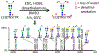
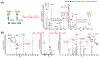
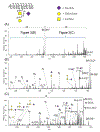


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

