Tracing a protein's folding pathway over evolutionary time using ancestral sequence reconstruction and hydrogen exchange
- PMID: 30204082
- PMCID: PMC6158009
- DOI: 10.7554/eLife.38369
Tracing a protein's folding pathway over evolutionary time using ancestral sequence reconstruction and hydrogen exchange
Abstract
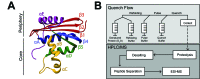
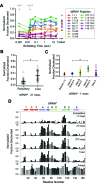
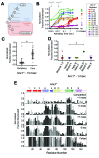
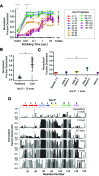
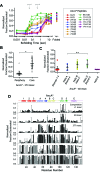
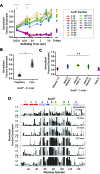
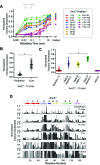
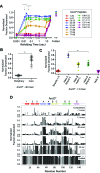
The conformations populated during protein folding have been studied for decades; yet, their evolutionary importance remains largely unexplored. Ancestral sequence reconstruction allows access to proteins across evolutionary time, and new methods such as pulsed-labeling hydrogen exchange coupled with mass spectrometry allow determination of folding intermediate structures at near amino-acid resolution. Here, we combine these techniques to monitor the folding of the ribonuclease H family along the evolutionary lineages of T. thermophilus and E. coli RNase H. All homologs and ancestral proteins studied populate a similar folding intermediate despite being separated by billions of years of evolution. Even though this conformation is conserved, the pathway leading to it has diverged over evolutionary time, and rational mutations can alter this trajectory. Our results demonstrate that evolutionary processes can affect the energy landscape to preserve or alter specific features of a protein's folding pathway.
Keywords: E. coli; ancestral sequence reconstruction; biochemistry; chemical biology; hydrogen exchange; mass spectrometry; molecular biophysics; protein evolution; protein folding; structural biology.
© 2018, Lim et al.
Conflict of interest statement
SL, EB, SM No competing interests declared
Figures





Similar articles
-
Evolutionary trend toward kinetic stability in the folding trajectory of RNases H.Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):13045-13050. doi: 10.1073/pnas.1611781113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799545 Free PMC article.
-
The burst-phase folding intermediate of ribonuclease H changes conformation over evolutionary history.Biopolymers. 2018 Aug;109(8):e23086. doi: 10.1002/bip.23086. Epub 2017 Nov 20. Biopolymers. 2018. PMID: 29152711 Free PMC article.
-
Comparison of the folding processes of T. thermophilus and E. coli ribonucleases H.J Mol Biol. 2002 Feb 15;316(2):327-40. doi: 10.1006/jmbi.2001.5346. J Mol Biol. 2002. PMID: 11851342
-
Protein folding intermediates and pathways studied by hydrogen exchange.Annu Rev Biophys Biomol Struct. 2000;29:213-38. doi: 10.1146/annurev.biophys.29.1.213. Annu Rev Biophys Biomol Struct. 2000. PMID: 10940248 Review.
-
Touring the landscapes: partially folded proteins examined by hydrogen exchange.Structure. 1997 Jul 15;5(7):859-63. doi: 10.1016/s0969-2126(97)00240-2. Structure. 1997. PMID: 9261079 Review.
Cited by
-
Ancestral Reconstruction and the Evolution of Protein Energy Landscapes.Annu Rev Biophys. 2024 Jul;53(1):127-146. doi: 10.1146/annurev-biophys-030722-125440. Epub 2024 Jun 28. Annu Rev Biophys. 2024. PMID: 38134334 Free PMC article. Review.
-
The Ensemble Basis of Allostery and Function: Insights from Models of Local Unfolding.J Mol Biol. 2025 Jun 9:169287. doi: 10.1016/j.jmb.2025.169287. Online ahead of print. J Mol Biol. 2025. PMID: 40499749 Review.
-
Opening and closing of a cryptic pocket in VP35 toggles it between two different RNA-binding modes.Elife. 2025 Sep 2;14:RP104514. doi: 10.7554/eLife.104514. Elife. 2025. PMID: 40892042 Free PMC article.
-
Opening of a cryptic pocket in β-lactamase increases penicillinase activity.Proc Natl Acad Sci U S A. 2021 Nov 23;118(47):e2106473118. doi: 10.1073/pnas.2106473118. Proc Natl Acad Sci U S A. 2021. PMID: 34799442 Free PMC article.
-
Evolution, folding, and design of TIM barrels and related proteins.Curr Opin Struct Biol. 2021 Jun;68:94-104. doi: 10.1016/j.sbi.2020.12.007. Epub 2021 Jan 13. Curr Opin Struct Biol. 2021. PMID: 33453500 Free PMC article. Review.
References
-
- Ahn M, Hagan CL, Bernardo-Gancedo A, De Genst E, Newby FN, Christodoulou J, Dhulesia A, Dumoulin M, Robinson CV, Dobson CM, Kumita JR. The significance of the location of mutations for the Native-State dynamics of human lysozyme. Biophysical Journal. 2016;111:2358–2367. doi: 10.1016/j.bpj.2016.10.028. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

