Understanding the role of glucose regulated protein 170 (GRP170) as a nucleotide exchange factor through molecular simulations
- PMID: 30205291
- PMCID: PMC6197907
- DOI: 10.1016/j.jmgm.2018.09.001
Understanding the role of glucose regulated protein 170 (GRP170) as a nucleotide exchange factor through molecular simulations
Abstract
Glucose Regulated Protein 170 (GRP170), also called Oxygen Regulated Protein 150 (ORP150), is a major molecular chaperone resident in the endoplasmic reticulum (ER). It belongs to the heat shock protein (HSP70) super family and can be induced by conditions such as hypoxia, ischemia and interferences in calcium homeostasis. It was recently reported that GRP170 may act as a nucleotide exchange factor (NEF) for GRP78 or binding immunoglobulin protein (BiP), and the ER canonical HSP70. However, little is known about the mechanism underlying its NEF activity. In this study, two homology models of GRP170 were constructed based on the X-ray crystal structures of ADP and ATP bound HSP110, a cytosolic homolog of GRP170, in order to characterize the differences in the binding modes of both ligands. It was observed that the differences in the binding modes of ADP and ATP led to a conformation change in the substrate binding domain which could potentially influence the binding of its substrates such as BiP. Our findings help understand the effect of nucleotide binding on the function of this chaperone protein as a NEF as well as the structural differences between GRP170 and its family members.
Keywords: Docking; GRP170; Homology modeling; Nucleotide binding; Nucleotide exchange factor (NEF); ORP150.
Published by Elsevier Inc.
Conflict of interest statement
Figures






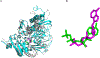
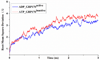

Similar articles
-
The endoplasmic reticulum Grp170 acts as a nucleotide exchange factor of Hsp70 via a mechanism similar to that of the cytosolic Hsp110.J Biol Chem. 2010 Apr 16;285(16):12445-53. doi: 10.1074/jbc.M109.096735. Epub 2010 Feb 20. J Biol Chem. 2010. PMID: 20177057 Free PMC article.
-
The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s.Cell Stress Chaperones. 2000 Oct;5(4):276-90. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. Cell Stress Chaperones. 2000. PMID: 11048651 Free PMC article. Review.
-
The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s.J Biol Chem. 2014 Jan 31;289(5):2899-907. doi: 10.1074/jbc.M113.507491. Epub 2013 Dec 10. J Biol Chem. 2014. PMID: 24327659 Free PMC article.
-
Nucleotide Exchange Factors for Hsp70 Molecular Chaperones: GrpE, Hsp110/Grp170, HspBP1/Sil1, and BAG Domain Proteins.Subcell Biochem. 2023;101:1-39. doi: 10.1007/978-3-031-14740-1_1. Subcell Biochem. 2023. PMID: 36520302 Review.
-
The 170 kDa glucose regulated stress protein is a large HSP70-, HSP110-like protein of the endoplasmic reticulum.FEBS Lett. 1996 Feb 12;380(1-2):68-72. doi: 10.1016/0014-5793(96)00011-7. FEBS Lett. 1996. PMID: 8603749
Cited by
-
The Essential Functions of Molecular Chaperones and Folding Enzymes in Maintaining Endoplasmic Reticulum Homeostasis.J Mol Biol. 2024 Jul 15;436(14):168418. doi: 10.1016/j.jmb.2023.168418. Epub 2023 Dec 22. J Mol Biol. 2024. PMID: 38143019 Free PMC article. Review.
-
Biological Function of HYOU1 in Tumors and Other Diseases.Onco Targets Ther. 2021 Mar 5;14:1727-1735. doi: 10.2147/OTT.S297332. eCollection 2021. Onco Targets Ther. 2021. PMID: 33707955 Free PMC article. Review.
-
Biochemical and biophysical characterization of Plasmodium falciparum glucose regulated protein 170.Sci Rep. 2025 Jul 29;15(1):27655. doi: 10.1038/s41598-025-98317-0. Sci Rep. 2025. PMID: 40730633 Free PMC article.
References
-
- Lee AS, Mammalian stress response: induction of the glucose-regulated protein family, Curr. Opin. Cell Biol 4 (1992) 267–273. - PubMed
-
- Pouysségur J, Shiu RP and Pastan I, Induction of two transformation-sensitive membrane polypeptides in normal fibroblasts by a block in glycoprotein synthesis or glucose deprivation, Cell 11 (1977) 941–947. - PubMed
-
- Gething M-J and Sambrook J, Protein folding in the cell, Nature 355 (1992) 33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

