Phosphorylation-Dependent Inhibition of Akt1
- PMID: 30205513
- PMCID: PMC6162393
- DOI: 10.3390/genes9090450
Phosphorylation-Dependent Inhibition of Akt1
Abstract
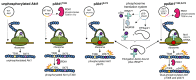
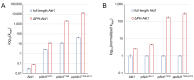
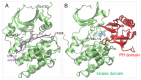
Protein kinase B (Akt1) is a proto-oncogene that is overactive in most cancers. Akt1 activation requires phosphorylation at Thr308; phosphorylation at Ser473 further enhances catalytic activity. Akt1 activity is also regulated via interactions between the kinase domain and the N-terminal auto-inhibitory pleckstrin homology (PH) domain. As it was previously difficult to produce Akt1 in site-specific phosphorylated forms, the contribution of each activating phosphorylation site to auto-inhibition was unknown. Using a combination of genetic code expansion and in vivo enzymatic phosphorylation, we produced Akt1 variants containing programmed phosphorylation to probe the interplay between Akt1 phosphorylation status and the auto-inhibitory function of the PH domain. Deletion of the PH domain increased the enzyme activity for all three phosphorylated Akt1 variants. For the doubly phosphorylated enzyme, deletion of the PH domain relieved auto-inhibition by 295-fold. We next found that phosphorylation at Ser473 provided resistance to chemical inhibition by Akti-1/2 inhibitor VIII. The Akti-1/2 inhibitor was most effective against pAkt1T308 and showed four-fold decreased potency with Akt1 variants phosphorylated at Ser473. The data highlight the need to design more potent Akt1 inhibitors that are effective against the doubly phosphorylated and most pathogenic form of Akt1.
Keywords: genetic code expansion; phosphoinositide dependent kinase 1; phosphoseryl-tRNA synthetase; protein kinase B; tRNASep.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Delivery of Active AKT1 to Human Cells.Cells. 2022 Nov 29;11(23):3834. doi: 10.3390/cells11233834. Cells. 2022. PMID: 36497091 Free PMC article.
-
Delivery of AKT1 phospho-forms to human cells reveals differential substrate selectivity.IUBMB Life. 2024 Sep;76(9):632-646. doi: 10.1002/iub.2826. Epub 2024 May 13. IUBMB Life. 2024. PMID: 38738523
-
Phosphorylation-dependent substrate selectivity of protein kinase B (AKT1).J Biol Chem. 2020 Jun 12;295(24):8120-8134. doi: 10.1074/jbc.RA119.012425. Epub 2020 Apr 29. J Biol Chem. 2020. PMID: 32350110 Free PMC article.
-
Genetic code expansion and live cell imaging reveal that Thr-308 phosphorylation is irreplaceable and sufficient for Akt1 activity.J Biol Chem. 2018 Jul 6;293(27):10744-10756. doi: 10.1074/jbc.RA118.002357. Epub 2018 May 17. J Biol Chem. 2018. PMID: 29773654 Free PMC article.
-
Phospho-Form Specific Substrates of Protein Kinase B (AKT1).Front Bioeng Biotechnol. 2021 Feb 3;8:619252. doi: 10.3389/fbioe.2020.619252. eCollection 2020. Front Bioeng Biotechnol. 2021. PMID: 33614606 Free PMC article.
Cited by
-
Delivery of Active AKT1 to Human Cells.Cells. 2022 Nov 29;11(23):3834. doi: 10.3390/cells11233834. Cells. 2022. PMID: 36497091 Free PMC article.
-
Effects of urolithin A on osteoclast differentiation induced by receptor activator of nuclear factor-κB ligand via bone morphogenic protein 2.Bioengineered. 2022 Mar;13(3):5064-5078. doi: 10.1080/21655979.2022.2036893. Bioengineered. 2022. PMID: 35164658 Free PMC article.
-
Isoliensinine induces cervical cancer cell cycle arrest and apoptosis by inhibiting the AKT/GSK3α pathway.Oncol Lett. 2022 Jan;23(1):8. doi: 10.3892/ol.2021.13126. Epub 2021 Nov 9. Oncol Lett. 2022. PMID: 34820007 Free PMC article.
-
MicroRNA-149 suppresses osteogenic differentiation of mesenchymal stem cells via inhibition of AKT1-dependent Twist1 phosphorylation.Cell Death Discov. 2022 Jan 10;8(1):2. doi: 10.1038/s41420-021-00618-6. Cell Death Discov. 2022. PMID: 35013126 Free PMC article.
-
Inhibition of lncRNA NFIA-AS1 Alleviates Abnormal Proliferation and Inflammation of Vascular Smooth Muscle Cells in Atherosclerosis by Regulating miR-125a-3p/AKT1 Axis.Int J Genomics. 2023 Apr 4;2023:8437898. doi: 10.1155/2023/8437898. eCollection 2023. Int J Genomics. 2023. PMID: 37056786 Free PMC article.
References
-
- Spencer A., Yoon S.S., Harrison S.J., Morris S.R., Smith D.A., Brigandi R.A., Gauvin J., Kumar R., Opalinska J.B., Chen C. The novel AKT inhibitor afuresertib shows favorable safety, pharmacokinetics, and clinical activity in multiple myeloma. Blood. 2014;124:2190–2195. doi: 10.1182/blood-2014-03-559963. - DOI - PMC - PubMed
-
- Antonelli M., Massimino M., Morra I., Garre M.L., Gardiman M.P., Buttarelli F.R., Arcella A., Giangaspero F. Expression of pERK and pAKT in pediatric high grade astrocytomas: Correlation with YKL40 and prognostic significance. Neuropathology. 2012;32:133–138. doi: 10.1111/j.1440-1789.2011.01252.x. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

