Targeting Amyloid Aggregation: An Overview of Strategies and Mechanisms
- PMID: 30205618
- PMCID: PMC6164555
- DOI: 10.3390/ijms19092677
Targeting Amyloid Aggregation: An Overview of Strategies and Mechanisms
Abstract
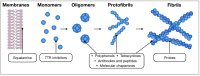
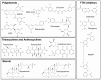
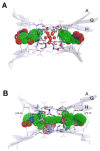
Amyloids result from the aggregation of a set of diverse proteins, due to either specific mutations or promoting intra- or extra-cellular conditions. Structurally, they are rich in intermolecular β-sheets and are the causative agents of several diseases, both neurodegenerative and systemic. It is believed that the most toxic species are small aggregates, referred to as oligomers, rather than the final fibrillar assemblies. Their mechanisms of toxicity are mostly mediated by aberrant interactions with the cell membranes, with resulting derangement of membrane-related functions. Much effort is being exerted in the search for natural antiamyloid agents, and/or in the development of synthetic molecules. Actually, it is well documented that the prevention of amyloid aggregation results in several cytoprotective effects. Here, we portray the state of the art in the field. Several natural compounds are effective antiamyloid agents, notably tetracyclines and polyphenols. They are generally non-specific, as documented by their partially overlapping mechanisms and the capability to interfere with the aggregation of several unrelated proteins. Among rationally designed molecules, we mention the prominent examples of β-breakers peptides, whole antibodies and fragments thereof, and the special case of drugs with contrasting transthyretin aggregation. In this framework, we stress the pivotal role of the computational approaches. When combined with biophysical methods, in several cases they have helped clarify in detail the protein/drug modes of interaction, which makes it plausible that more effective drugs will be developed in the future.
Keywords: amyloid diseases; biocomputing; drug design; natural antiamyloids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

