Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family
- PMID: 30206120
- PMCID: PMC6204895
- DOI: 10.1074/jbc.RA117.000733
Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family
Abstract
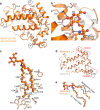
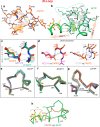
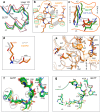
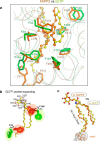
The glycolipid transfer protein (GLTP) fold defines a superfamily of eukaryotic proteins that selectively transport sphingolipids (SLs) between membranes. However, the mechanisms determining the protein selectivity for specific glycosphingolipids (GSLs) are unclear. Here, we report the crystal structure of the GLTP homology (GLTPH) domain of human 4-phosphate adaptor protein 2 (FAPP2) bound with N-oleoyl-galactosylceramide. Using this domain, FAPP2 transports glucosylceramide from its cis-Golgi synthesis site to the trans-Golgi for conversion into complex GSLs. The FAPP2-GLTPH structure revealed an element, termed the ID loop, that controls specificity in the GLTP family. We found that, in accordance with FAPP2 preference for simple GSLs, the ID loop protrudes from behind the SL headgroup-recognition center to mitigate binding by complex GSLs. Mutational analyses including GLTP and FAPP2 chimeras with swapped ID loops supported the proposed restrictive role of the FAPP2 ID loop in GSL selectivity. Comparative analysis revealed distinctly designed ID loops in each GLTP family member. This analysis also disclosed a conserved H-bond triplet that "clasps" both ID-loop ends together to promote structural autonomy and rigidity. The findings indicated that various ID loops work in concert with conserved recognition centers to create different specificities among family members. We also observed four bulky, conserved hydrophobic residues involved in "sensor-like" interactions with lipid chains in protein hydrophobic pockets and FF motifs in GLTP and FAPP2, well-positioned to provide acyl chain-dependent SL selectivity for the hydrophobic pockets. In summary, our study provides mechanistic insights into sphingolipid recognition by the GLTP fold and uncovers the elements involved in this recognition.
Keywords: 4-phosphate-adaptor-protein-2; FAPP2; X-ray crystallography; lipid-protein interaction; nonvesicular lipid transport; protein complex; protein family; protein structure; sphingolipid; transport.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- D'Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C. C., van der Spoel A. C., Platt F. M., Hannun Y. A., Polishchuk R., Mattjus P., and De Matteis M. A. (2007) Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature 449, 62–67 - PubMed
-
- Kamlekar R. K., Simanshu D. K., Gao Y. G., Kenoth R., Pike H. M., Prendergast F. G., Malinina L., Molotkovsky J. G., Venyaminov S. Y., Patel D. J., and Brown R. E. (2013) The glycolipid transfer protein (GLTP) domain of phosphoinositol 4-phosphate adaptor protein-2 (FAPP2): structure drives preference for simple neutral glycosphingolipids. Biochim. Biophys. Acta 1831, 417–427 - PMC - PubMed
-
- D'Angelo G., Uemura T., Chuang C. C., Polishchuk E., Santoro M., Ohvo-Rekila H., Sato T., Di Tullio G., Varriale A., D'Auria S., Daniele T., Capuani F., Johannes L., Mattjus P., Monti M., Pucci P., Williams R. L., Burke J. E., Platt F. M., Harada A., and De Matteis M. A. (2013) Vesicular and non-vesicular transport feed distinct glycosylation pathways in the Golgi. Nature 501, 116–120 - PubMed
-
- Mattjus P. (2009) Glycolipid transfer proteins and membrane interaction. Biochim. Biophys. Acta 1788, 267–272 - PubMed
-
- Abe A., Yamada K., and Sasaki T. (1982) A protein purified from pig brain accelerates the inter-membranous translocation of mono- and dihexosylceramides, but not the translocation of phospholipids. Biochem. Biophys. Res. Commun. 104, 1386–1393 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

